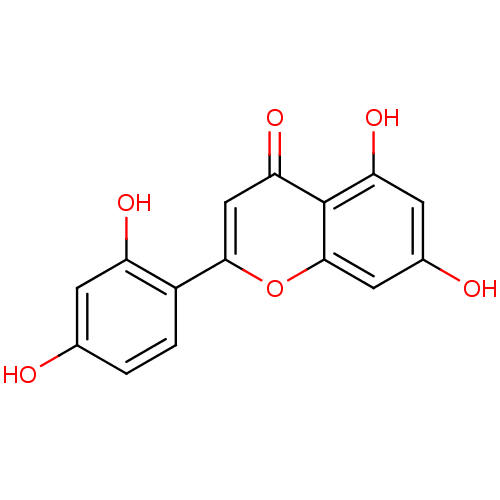
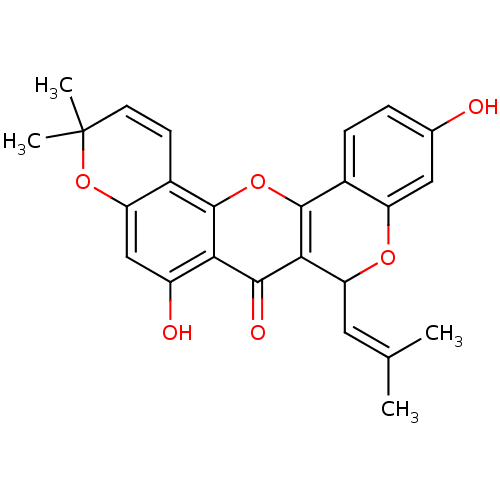
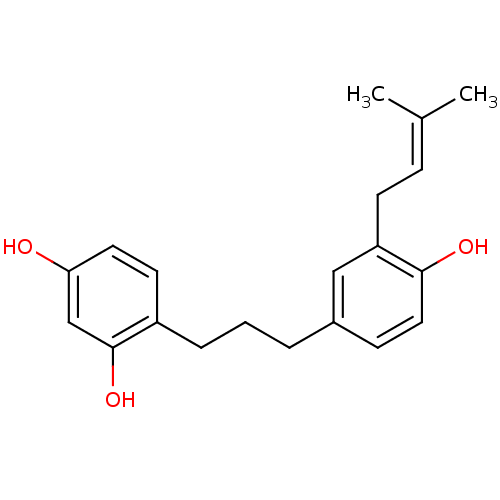
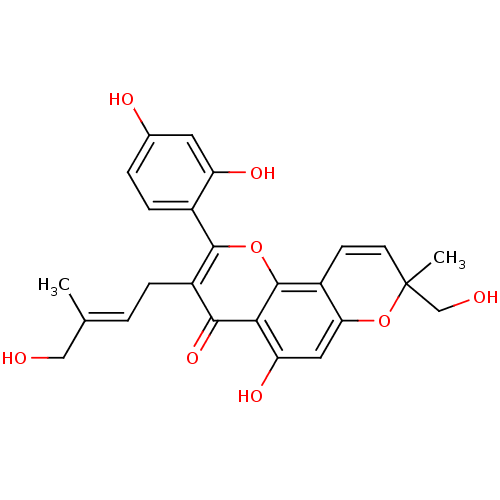
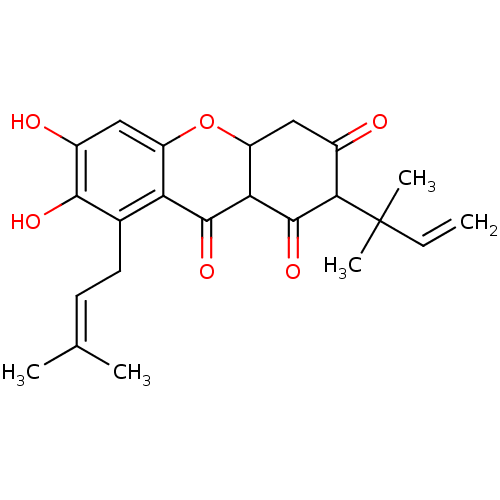
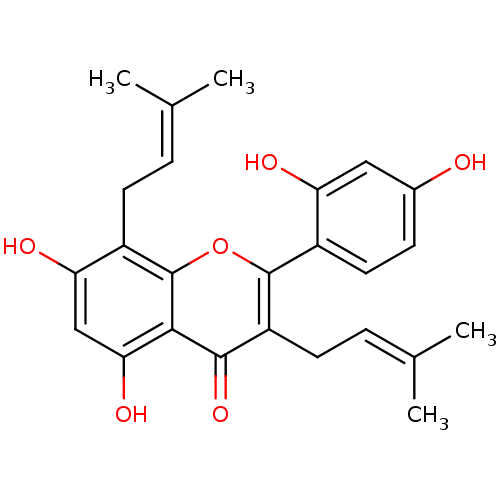
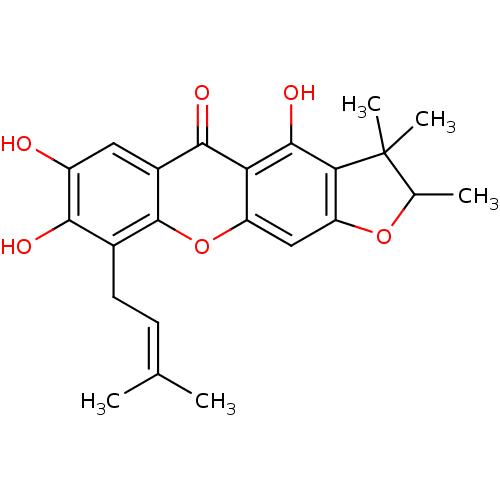
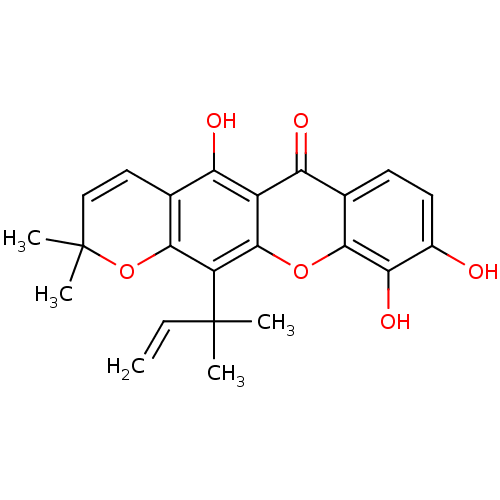
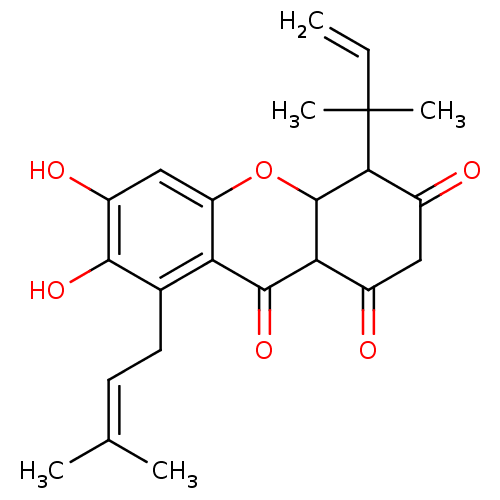
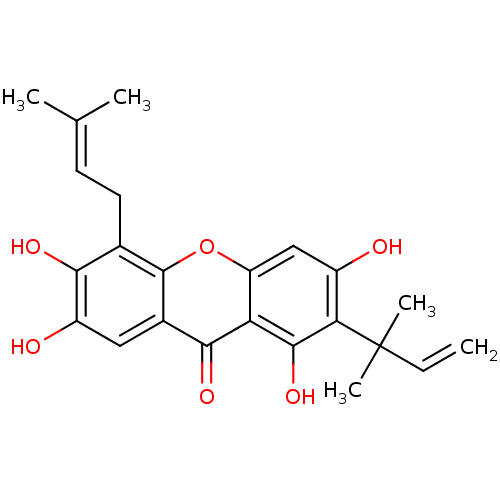
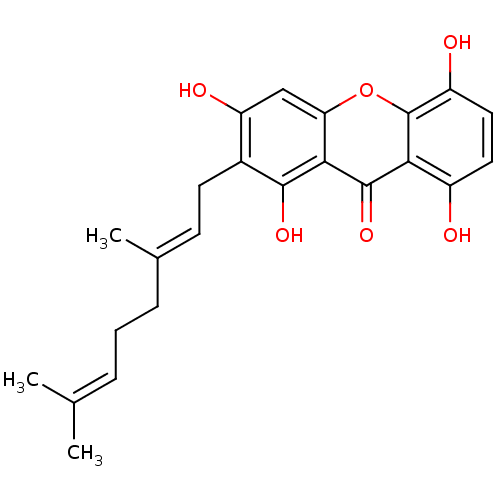
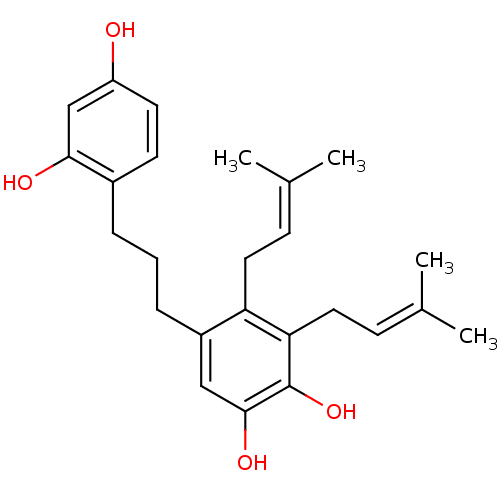
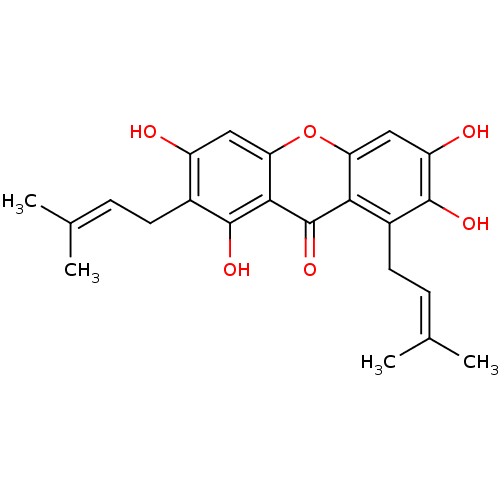
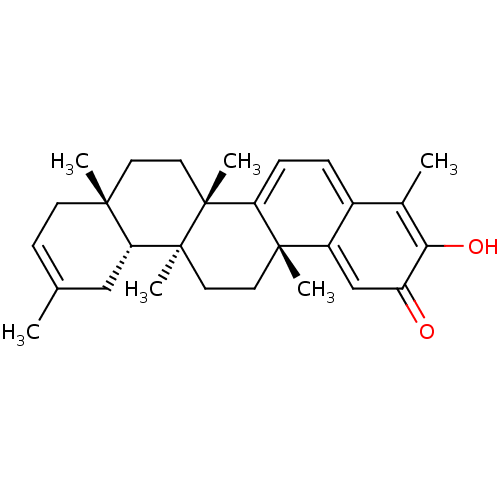
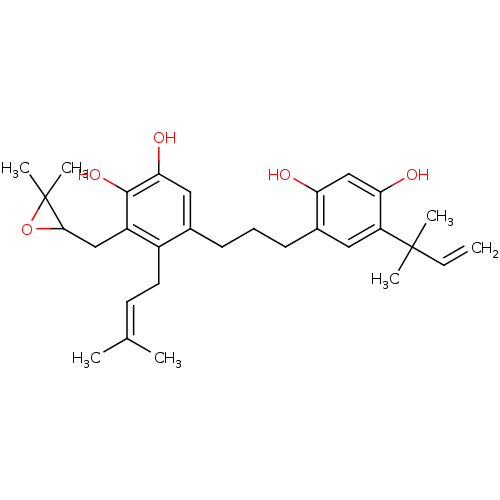
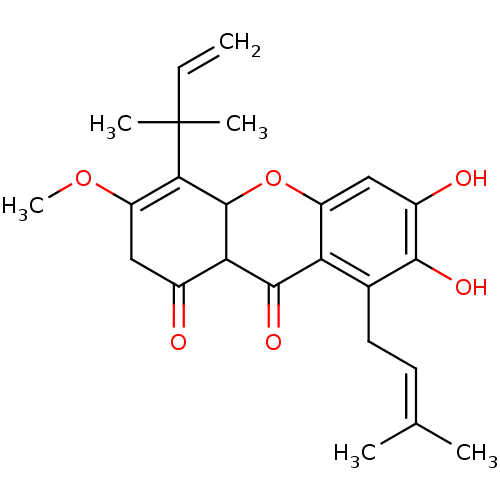
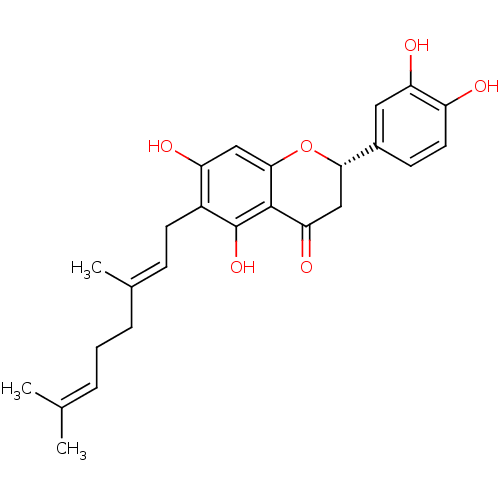
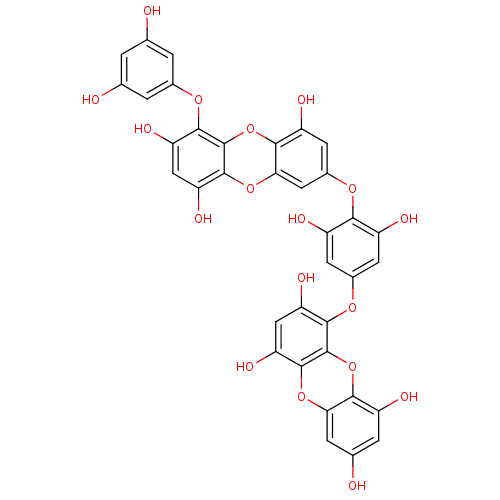
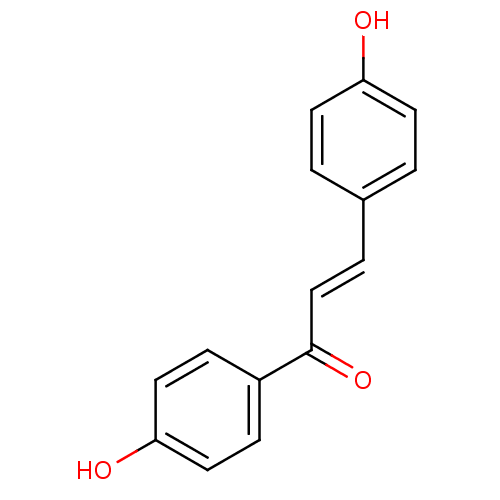
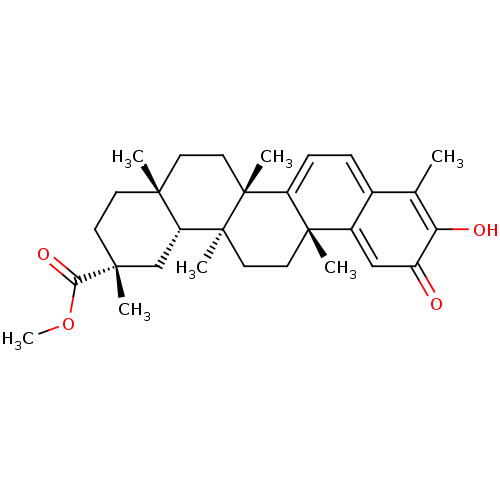
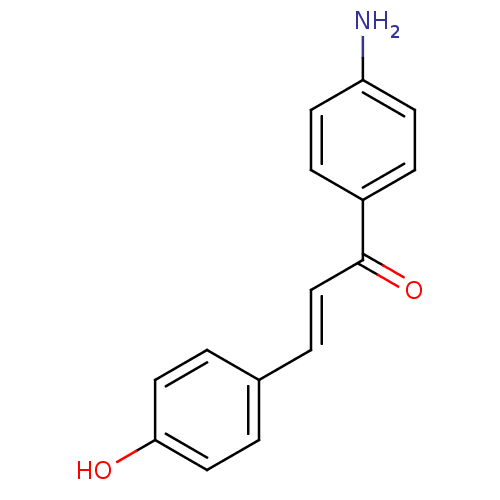
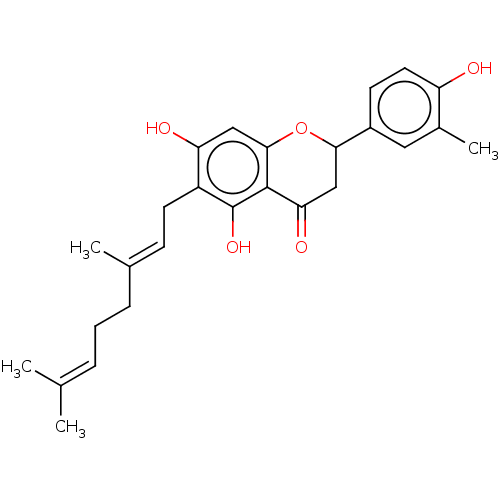
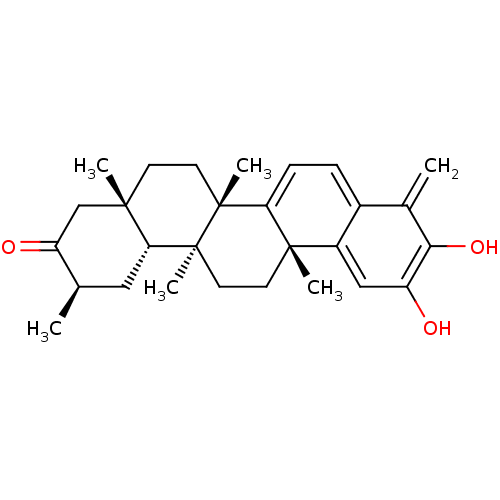
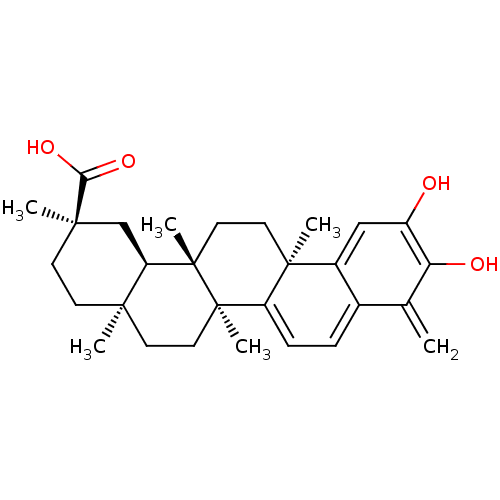
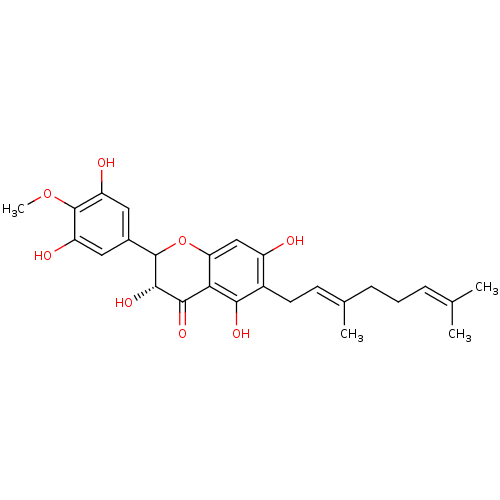
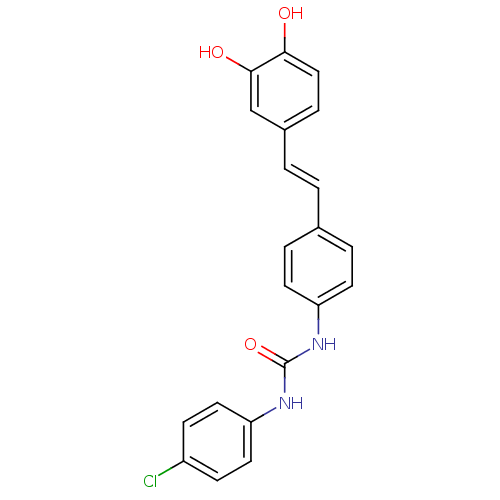
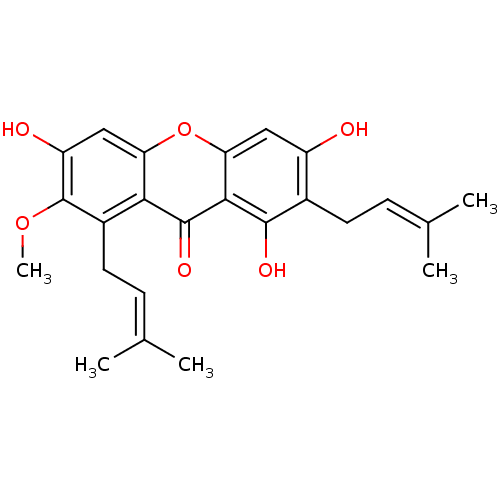
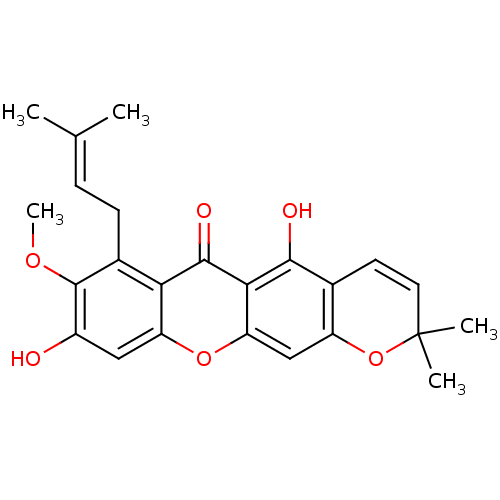
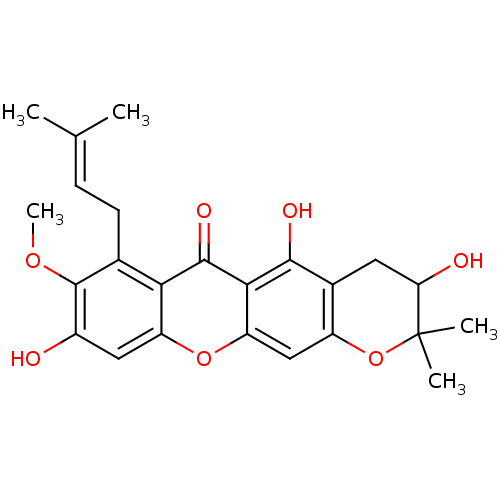
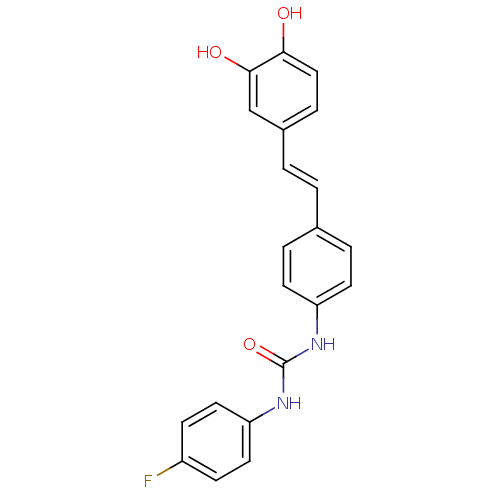
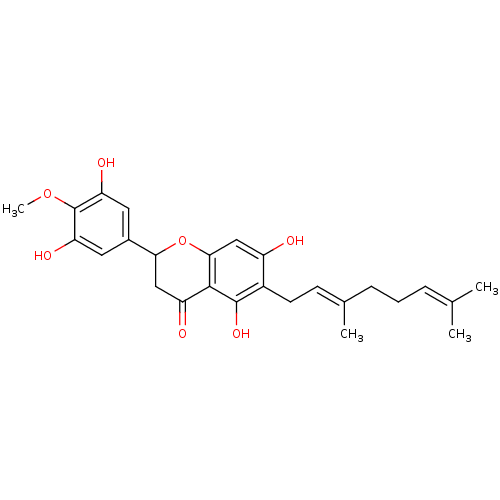
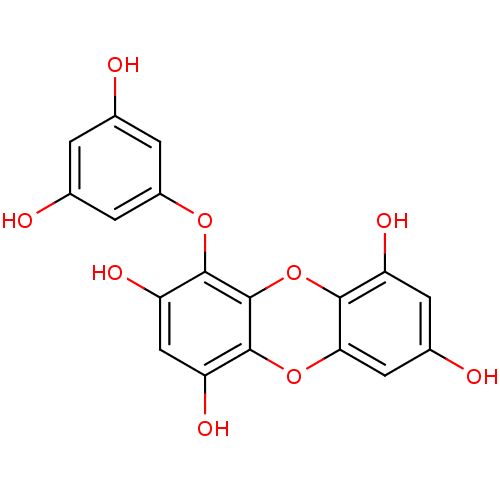
Affinity DataKi: 0.610nM ΔG°: -53.5kJ/moleT: 2°CAssay Description:Mushroom tyrosinase using either L-DOPA or L-tyrosine as substrate. In spectrophotometric experiments, enzyme activity was monitored by dopachrome f...More data for this Ligand-Target Pair
Affinity DataKi: 46.4nM ΔG°: -42.6kJ/moleT: 2°CAssay Description:Mushroom tyrosinase using either L-DOPA or L-tyrosine as substrate. In spectrophotometric experiments, enzyme activity was monitored by dopachrome f...More data for this Ligand-Target Pair
Affinity DataKi: 48.5nMAssay Description:Inhibition of mushroom tyrosinase by kinetic based assayMore data for this Ligand-Target Pair
Affinity DataKi: 49.2nM ΔG°: -42.4kJ/moleT: 2°CAssay Description:Mushroom tyrosinase using either L-DOPA or L-tyrosine as substrate. In spectrophotometric experiments, enzyme activity was monitored by dopachrome f...More data for this Ligand-Target Pair
TargetSialidase(Clostridium perfringens)
Graduate School Of Gyeongsang National University
Curated by ChEMBL
Graduate School Of Gyeongsang National University
Curated by ChEMBL
Affinity DataKi: 58nMAssay Description:Competitive inhibition of Clostridium perfringens neuraminidase by Lineweaver-Burke plot and Dixon plotMore data for this Ligand-Target Pair
Affinity DataKi: 80.5nM ΔG°: -41.2kJ/moleT: 2°CAssay Description:Mushroom tyrosinase using either L-DOPA or L-tyrosine as substrate. In spectrophotometric experiments, enzyme activity was monitored by dopachrome f...More data for this Ligand-Target Pair
TargetSialidase(Clostridium perfringens)
Graduate School Of Gyeongsang National University
Curated by ChEMBL
Graduate School Of Gyeongsang National University
Curated by ChEMBL
Affinity DataKi: 98nMAssay Description:Competitive inhibition of Clostridium perfringens neuraminidase by Lineweaver-Burke plot and Dixon plotMore data for this Ligand-Target Pair
TargetSialidase(Clostridium perfringens)
Graduate School Of Gyeongsang National University
Curated by ChEMBL
Graduate School Of Gyeongsang National University
Curated by ChEMBL
Affinity DataKi: 103nMAssay Description:Competitive inhibition of Clostridium perfringens neuraminidase by Lineweaver-Burke plot and Dixon plotMore data for this Ligand-Target Pair
TargetSialidase(Clostridium perfringens)
Graduate School Of Gyeongsang National University
Curated by ChEMBL
Graduate School Of Gyeongsang National University
Curated by ChEMBL
Affinity DataKi: 127nMAssay Description:Apparent binding affinity at Clostridium perfringens neuraminidase by fluorimetryMore data for this Ligand-Target Pair
TargetSialidase(Clostridium perfringens)
Graduate School Of Gyeongsang National University
Curated by ChEMBL
Graduate School Of Gyeongsang National University
Curated by ChEMBL
Affinity DataKi: 136nMAssay Description:Competitive inhibition of Clostridium perfringens neuraminidase by Lineweaver-Burke plot and Dixon plotMore data for this Ligand-Target Pair
TargetSialidase(Clostridium perfringens)
Graduate School Of Gyeongsang National University
Curated by ChEMBL
Graduate School Of Gyeongsang National University
Curated by ChEMBL
Affinity DataKi: 138nMAssay Description:Competitive inhibition of Clostridium perfringens neuraminidase by Lineweaver-Burke plot and Dixon plotMore data for this Ligand-Target Pair
TargetSialidase(Clostridium perfringens)
Graduate School Of Gyeongsang National University
Curated by ChEMBL
Graduate School Of Gyeongsang National University
Curated by ChEMBL
Affinity DataKi: 143nMAssay Description:Competitive inhibition of Clostridium perfringens neuraminidase by Lineweaver-Burke plot and Dixon plotMore data for this Ligand-Target Pair
TargetSialidase(Clostridium perfringens)
Graduate School Of Gyeongsang National University
Curated by ChEMBL
Graduate School Of Gyeongsang National University
Curated by ChEMBL
Affinity DataKi: 150nMAssay Description:Competitive inhibition of Clostridium perfringens neuraminidase by fluorometryMore data for this Ligand-Target Pair
Affinity DataKi: 181nMAssay Description:Inhibition of mushroom tyrosinase by kinetic based assayMore data for this Ligand-Target Pair
TargetSialidase(Clostridium perfringens)
Graduate School Of Gyeongsang National University
Curated by ChEMBL
Graduate School Of Gyeongsang National University
Curated by ChEMBL
Affinity DataKi: 214nMAssay Description:Apparent binding affinity at Clostridium perfringens neuraminidase by fluorimetryMore data for this Ligand-Target Pair
Affinity DataKi: 230nMAssay Description:Inhibition of monophenolase activity of mushroom tyrosinase using as L-tyrosine substrate by Lineweaver-Burk plot based kinetic assayMore data for this Ligand-Target Pair
Affinity DataKi: 290nMAssay Description:Inhibition of diphenolase activity of mushroom tyrosinase using as L-DOPA substrate by Lineweaver-Burk plot based kinetic assayMore data for this Ligand-Target Pair
Affinity DataKi: 410nMAssay Description:Inhibition of monophenolase activity of mushroom tyrosinase using as L-tyrosine substrate by Lineweaver-Burk plot based kinetic assayMore data for this Ligand-Target Pair
TargetSialidase(Clostridium perfringens)
Graduate School Of Gyeongsang National University
Curated by ChEMBL
Graduate School Of Gyeongsang National University
Curated by ChEMBL
Affinity DataKi: 436nMAssay Description:Apparent binding affinity at Clostridium perfringens neuraminidase by fluorimetryMore data for this Ligand-Target Pair
TargetSialidase(Clostridium perfringens)
Graduate School Of Gyeongsang National University
Curated by ChEMBL
Graduate School Of Gyeongsang National University
Curated by ChEMBL
Affinity DataKi: 477nMAssay Description:Apparent binding affinity at Clostridium perfringens neuraminidase by fluorimetryMore data for this Ligand-Target Pair
TargetSialidase(Clostridium perfringens)
Graduate School Of Gyeongsang National University
Curated by ChEMBL
Graduate School Of Gyeongsang National University
Curated by ChEMBL
Affinity DataKi: 510nMAssay Description:Apparent binding affinity at Clostridium perfringens neuraminidase by fluorimetryMore data for this Ligand-Target Pair
TargetSialidase(Clostridium perfringens)
Graduate School Of Gyeongsang National University
Curated by ChEMBL
Graduate School Of Gyeongsang National University
Curated by ChEMBL
Affinity DataKi: 568nMAssay Description:Apparent binding affinity at Clostridium perfringens neuraminidase by fluorimetryMore data for this Ligand-Target Pair
Affinity DataKi: 770nMAssay Description:Inhibition of diphenolase activity of mushroom tyrosinase using as L-DOPA substrate by Lineweaver-Burk plot based kinetic assayMore data for this Ligand-Target Pair
TargetSialidase(Clostridium perfringens)
Graduate School Of Gyeongsang National University
Curated by ChEMBL
Graduate School Of Gyeongsang National University
Curated by ChEMBL
Affinity DataKi: 800nMAssay Description:Competitive inhibition of Clostridium perfringens neuraminidase by fluorometryMore data for this Ligand-Target Pair
TargetReplicase polyprotein 1ab(Human SARS coronavirus (SARS-CoV) (Severe acute re...)
Korea Research Institute Of Bioscience And Biotechnology
Curated by ChEMBL
Korea Research Institute Of Bioscience And Biotechnology
Curated by ChEMBL
Affinity DataKi: 800nMAssay Description:Inhibition of 3C-like protease of SARS coronavirus assessed as concentration of FRET peptide for 60 mins by dixon plotMore data for this Ligand-Target Pair
Affinity DataKi: 870nMAssay Description:Inhibition of mushroom tyrosinase by kinetic based assayMore data for this Ligand-Target Pair
TargetSialidase(Clostridium perfringens)
Graduate School Of Gyeongsang National University
Curated by ChEMBL
Graduate School Of Gyeongsang National University
Curated by ChEMBL
Affinity DataKi: 950nMAssay Description:Competitive inhibition of Clostridium perfringens neuraminidase by Lineweaver-Burke plot and Dixon plotMore data for this Ligand-Target Pair
TargetCarboxylic ester hydrolase(Equus caballus (Horse))
Graduate School Of Gyeongsang National University
Curated by ChEMBL
Graduate School Of Gyeongsang National University
Curated by ChEMBL
Affinity DataKi: 1.20E+3nMAssay Description:Mixed type inhibition of equine BChE using butyrylthiocholine iodide as substrate by Lineweaver-Burk double-reciprocal-plot and dixon plot analysisMore data for this Ligand-Target Pair
Affinity DataKi: 2.20E+3nMAssay Description:Noncompetitive inhibition of human recombinant BACE-1 by Dixon plot analysisMore data for this Ligand-Target Pair
TargetSialidase(Clostridium perfringens)
Graduate School Of Gyeongsang National University
Curated by ChEMBL
Graduate School Of Gyeongsang National University
Curated by ChEMBL
Affinity DataKi: 2.23E+3nMAssay Description:Apparent binding affinity at Clostridium perfringens neuraminidase by fluorimetryMore data for this Ligand-Target Pair
TargetReplicase polyprotein 1ab(Human SARS coronavirus (SARS-CoV) (Severe acute re...)
Korea Research Institute Of Bioscience And Biotechnology
Curated by ChEMBL
Korea Research Institute Of Bioscience And Biotechnology
Curated by ChEMBL
Affinity DataKi: 2.40E+3nMAssay Description:Competitive inhibition of C-terminal His6-tagged recombinant SARS coronavirus 3C-like protease trans-cleavage activity expressed in Escherichia coli ...More data for this Ligand-Target Pair
Affinity DataKi: 2.90E+3nMAssay Description:Competitive inhibition of monophenolase activity of mushroom tyrosinase using as L-tyrosine substrate by Lineweaver-Burk plot analysisMore data for this Ligand-Target Pair
TargetReplicase polyprotein 1ab(Human SARS coronavirus (SARS-CoV) (Severe acute re...)
Korea Research Institute Of Bioscience And Biotechnology
Curated by ChEMBL
Korea Research Institute Of Bioscience And Biotechnology
Curated by ChEMBL
Affinity DataKi: 3.10E+3nMAssay Description:Inhibition of 3C-like protease of SARS coronavirus assessed as concentration of FRET peptide for 60 mins by dixon plotMore data for this Ligand-Target Pair
Affinity DataKi: 3.20E+3nM IC50: 8.40E+3nMAssay Description:All enzymatic activities were determined by using the appropriate substrate (p-nitrophenyl-alpha-D-glucopyranoside, p-nitrophenyl-beta-D-gulcopyranos...More data for this Ligand-Target Pair
TargetSialidase(Clostridium perfringens)
Graduate School Of Gyeongsang National University
Curated by ChEMBL
Graduate School Of Gyeongsang National University
Curated by ChEMBL
Affinity DataKi: 3.30E+3nMAssay Description:Competitive inhibition of Clostridium perfringens neuraminidase by fluorometryMore data for this Ligand-Target Pair
TargetSialidase(Clostridium perfringens)
Graduate School Of Gyeongsang National University
Curated by ChEMBL
Graduate School Of Gyeongsang National University
Curated by ChEMBL
Affinity DataKi: 3.60E+3nMAssay Description:Competitive inhibition of Clostridium perfringens neuraminidase by fluorometryMore data for this Ligand-Target Pair
Affinity DataKi: 3.90E+3nMAssay Description:Competitive inhibition of monophenolase activity of mushroom tyrosinase using as L-tyrosine substrate by Lineweaver-Burk plot analysisMore data for this Ligand-Target Pair
TargetSialidase(Clostridium perfringens)
Industry-Academic Cooperation Foundation Gyeongsang National University
US Patent
Industry-Academic Cooperation Foundation Gyeongsang National University
US Patent
Affinity DataKi: 4.00E+3nMAssay Description:Specifically, in order to determine IC50 of the compounds for neuraminidase, 0.01 U/ml of neuraminidase (EC. 3.2.1.8, C. perfringens, SIGMA, N2876) a...More data for this Ligand-Target Pair
Ligand Info
TargetReplicase polyprotein 1ab(Human SARS coronavirus (SARS-CoV) (Severe acute re...)
Korea Research Institute Of Bioscience And Biotechnology
Curated by ChEMBL
Korea Research Institute Of Bioscience And Biotechnology
Curated by ChEMBL
Affinity DataKi: 4.00E+3nMAssay Description:Inhibition of 3C-like protease of SARS coronavirus assessed as concentration of FRET peptide for 60 mins by dixon plotMore data for this Ligand-Target Pair
TargetReplicase polyprotein 1ab(Human SARS coronavirus (SARS-CoV) (Severe acute re...)
Korea Research Institute Of Bioscience And Biotechnology
Curated by ChEMBL
Korea Research Institute Of Bioscience And Biotechnology
Curated by ChEMBL
Affinity DataKi: 4.20E+3nMAssay Description:Inhibition of 3C-like protease of SARS coronavirus assessed as concentration of FRET peptide for 60 mins by dixon plotMore data for this Ligand-Target Pair
TargetCarboxylic ester hydrolase(Equus caballus (Horse))
Graduate School Of Gyeongsang National University
Curated by ChEMBL
Graduate School Of Gyeongsang National University
Curated by ChEMBL
Affinity DataKi: 4.50E+3nMAssay Description:Mixed type inhibition of equine BChE using butyrylthiocholine iodide as substrate by Lineweaver-Burk double-reciprocal-plot and dixon plot analysisMore data for this Ligand-Target Pair
Affinity DataKi: 4.60E+3nM IC50: 1.43E+4nMAssay Description:All enzymatic activities were determined by using the appropriate substrate (p-nitrophenyl-alpha-D-glucopyranoside, p-nitrophenyl-beta-D-gulcopyranos...More data for this Ligand-Target Pair
TargetSialidase(Clostridium perfringens)
Graduate School Of Gyeongsang National University
Curated by ChEMBL
Graduate School Of Gyeongsang National University
Curated by ChEMBL
Affinity DataKi: 5.80E+3nMAssay Description:Competitive inhibition of Clostridium perfringens neuraminidase by fluorometryMore data for this Ligand-Target Pair
TargetSialidase(Clostridium perfringens)
Graduate School Of Gyeongsang National University
Curated by ChEMBL
Graduate School Of Gyeongsang National University
Curated by ChEMBL
Affinity DataKi: 5.90E+3nMAssay Description:Competitive inhibition of Clostridium perfringens neuraminidase by fluorometryMore data for this Ligand-Target Pair
TargetSialidase(Clostridium perfringens)
Graduate School Of Gyeongsang National University
Curated by ChEMBL
Graduate School Of Gyeongsang National University
Curated by ChEMBL
Affinity DataKi: 6.50E+3nMAssay Description:Competitive inhibition of Clostridium perfringens neuraminidase by fluorometryMore data for this Ligand-Target Pair
TargetSialidase(Clostridium perfringens)
Graduate School Of Gyeongsang National University
Curated by ChEMBL
Graduate School Of Gyeongsang National University
Curated by ChEMBL
Affinity DataKi: 6.80E+3nMAssay Description:Competitive inhibition of Clostridium perfringens neuraminidase by fluorometryMore data for this Ligand-Target Pair
Affinity DataKi: 7.20E+3nM IC50: 1.98E+4nMAssay Description:All enzymatic activities were determined by using the appropriate substrate (p-nitrophenyl-alpha-D-glucopyranoside, p-nitrophenyl-beta-D-gulcopyranos...More data for this Ligand-Target Pair
TargetCarboxylic ester hydrolase(Equus caballus (Horse))
Graduate School Of Gyeongsang National University
Curated by ChEMBL
Graduate School Of Gyeongsang National University
Curated by ChEMBL
Affinity DataKi: 7.60E+3nMAssay Description:Mixed type inhibition of equine BChE using butyrylthiocholine iodide as substrate by Lineweaver-Burk double-reciprocal-plot and dixon plot analysisMore data for this Ligand-Target Pair
TargetSialidase(Clostridium perfringens)
Graduate School Of Gyeongsang National University
Curated by ChEMBL
Graduate School Of Gyeongsang National University
Curated by ChEMBL
Affinity DataKi: 7.60E+3nMAssay Description:Competitive inhibition of Clostridium perfringens neuraminidase by fluorometryMore data for this Ligand-Target Pair
TargetReplicase polyprotein 1ab(Human SARS coronavirus (SARS-CoV) (Severe acute re...)
Korea Research Institute Of Bioscience And Biotechnology
Curated by ChEMBL
Korea Research Institute Of Bioscience And Biotechnology
Curated by ChEMBL
Affinity DataKi: 8.20E+3nMAssay Description:Competitive inhibition of C-terminal His6-tagged recombinant SARS coronavirus 3C-like protease trans-cleavage activity expressed in Escherichia coli ...More data for this Ligand-Target Pair