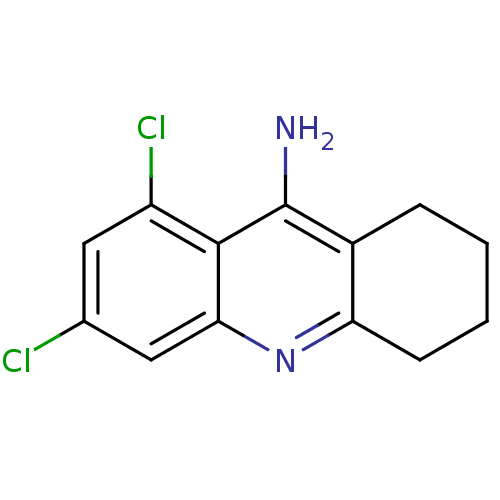
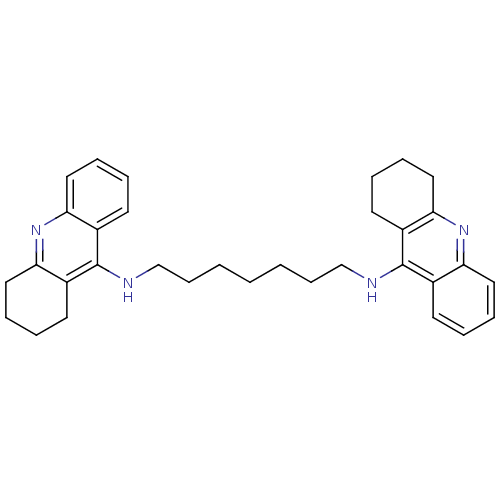
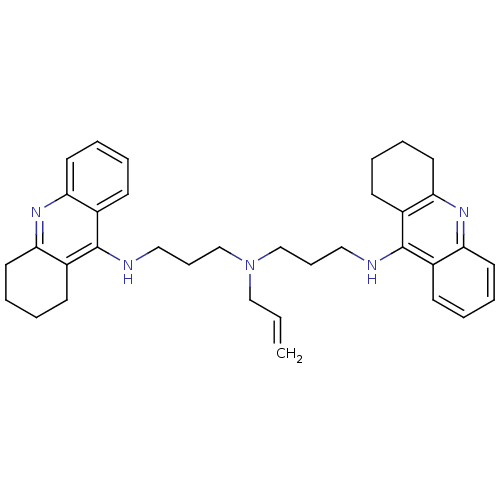
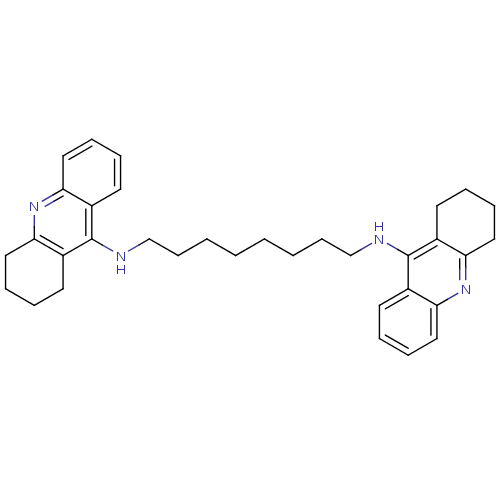
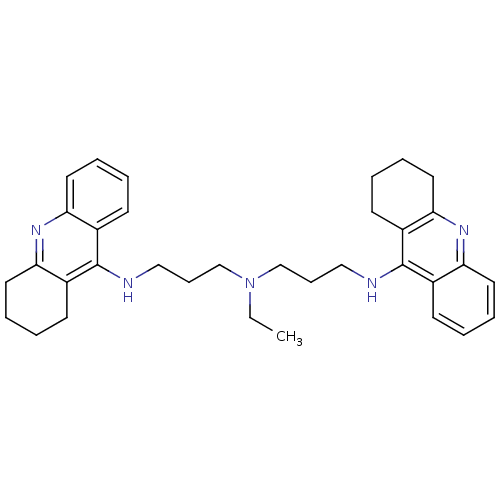
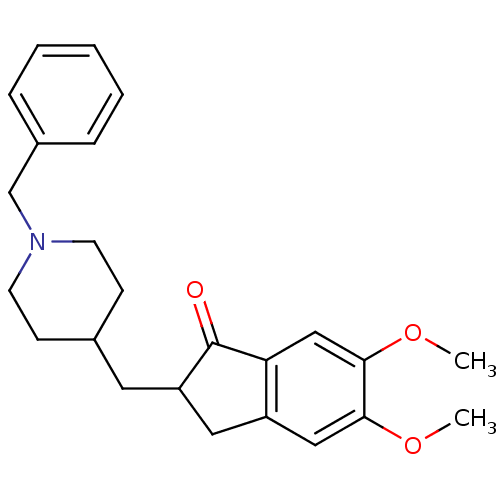
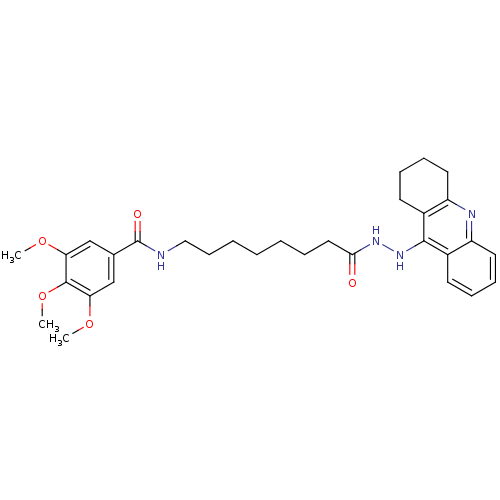
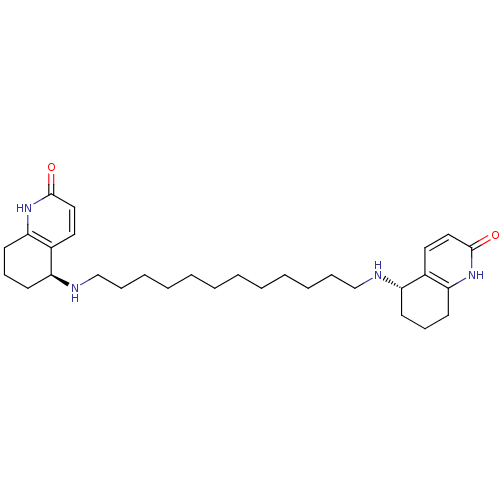
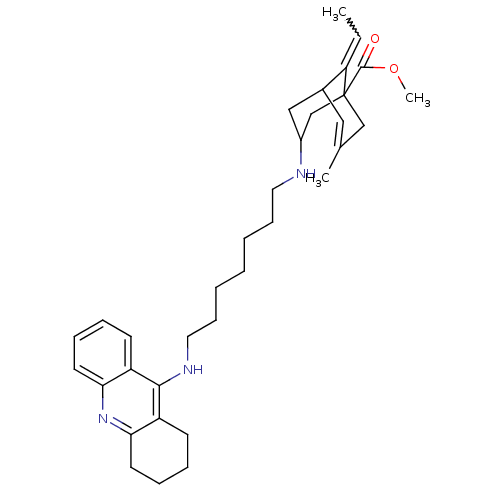
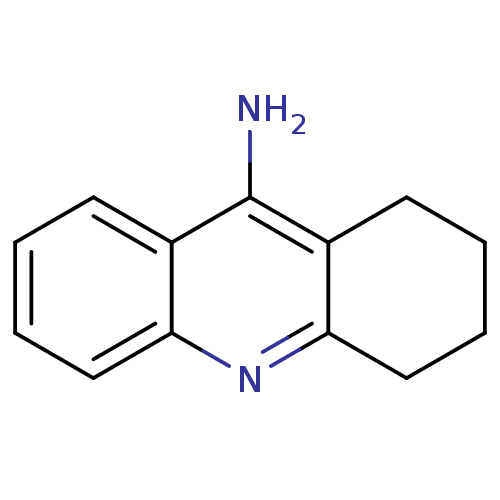
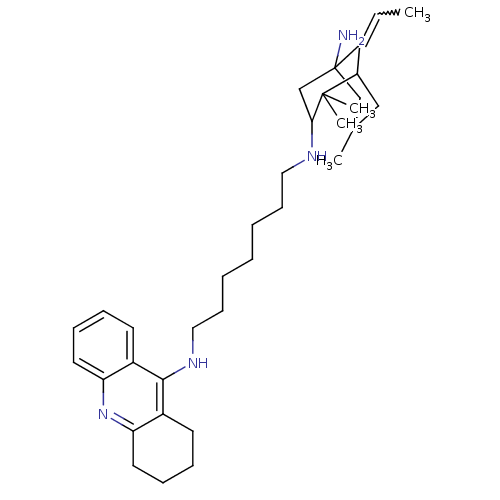
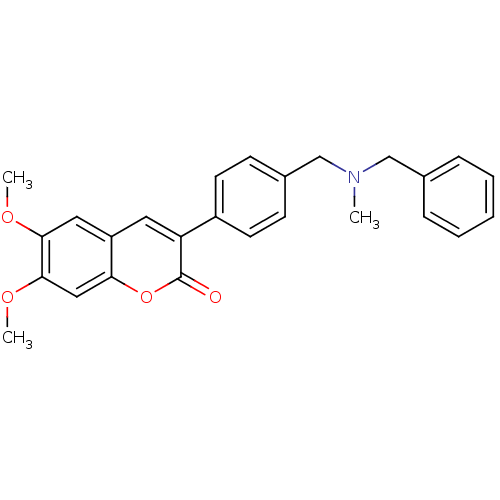
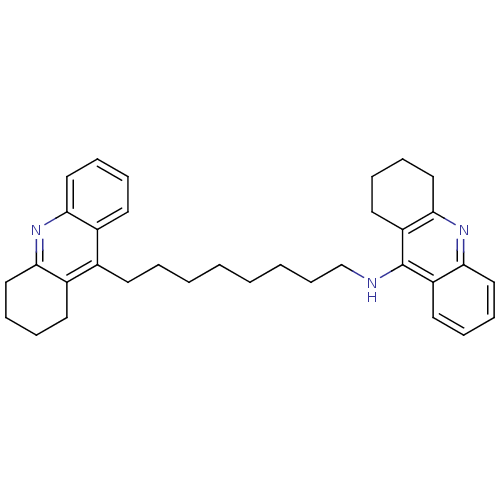
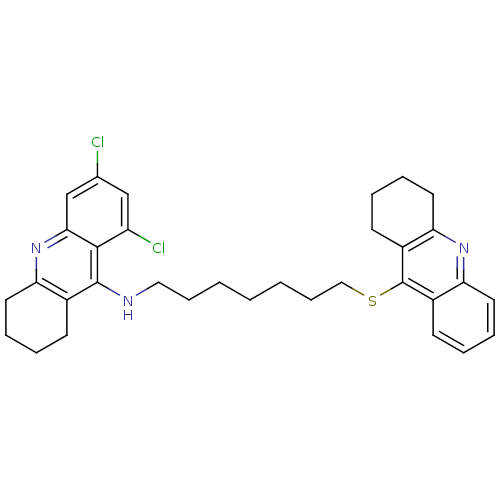
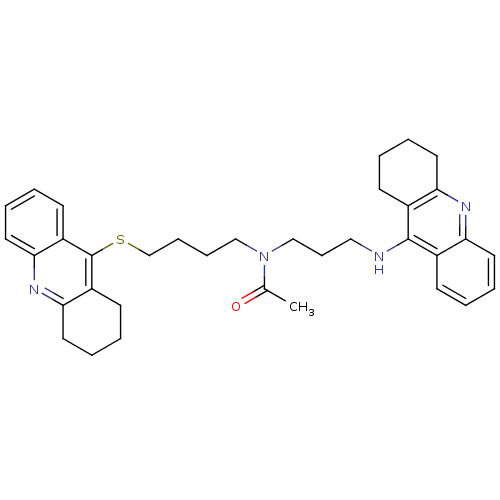
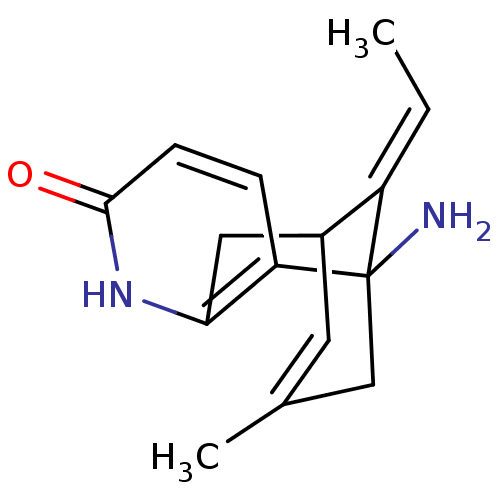
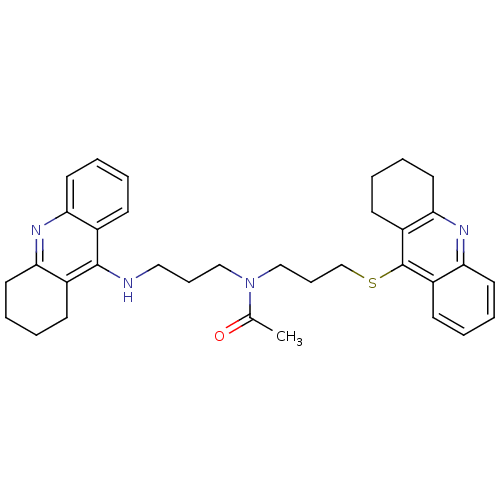
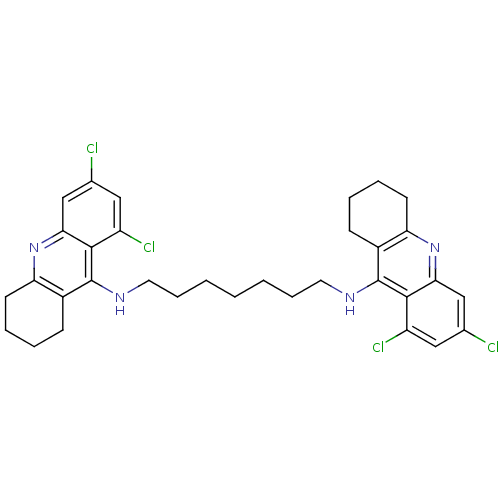
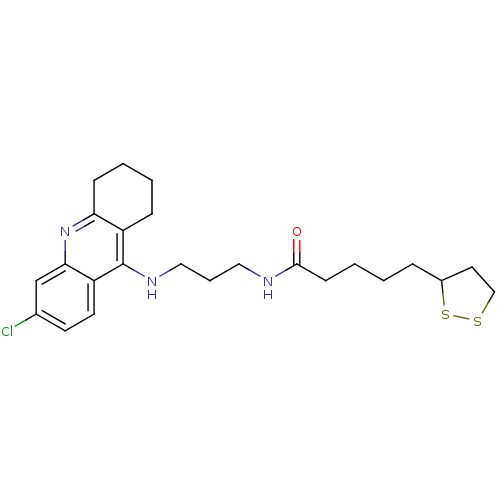
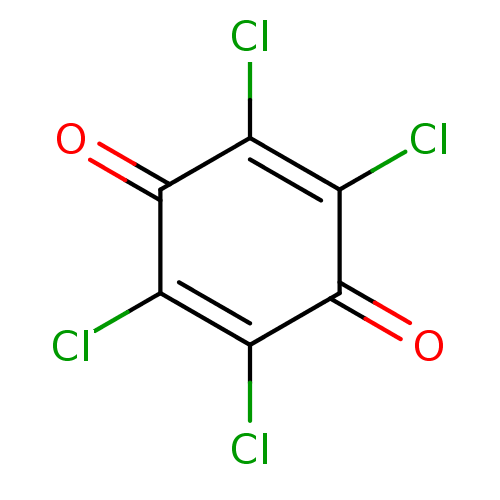
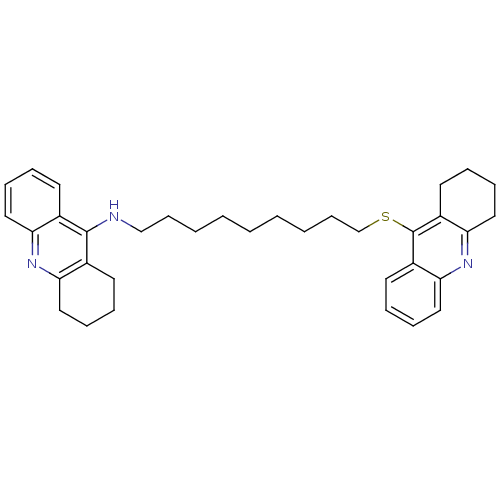
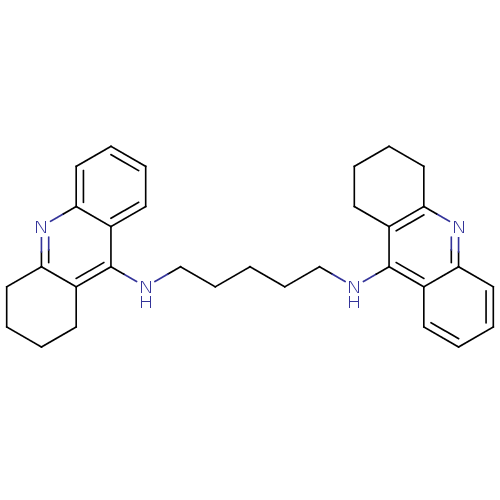
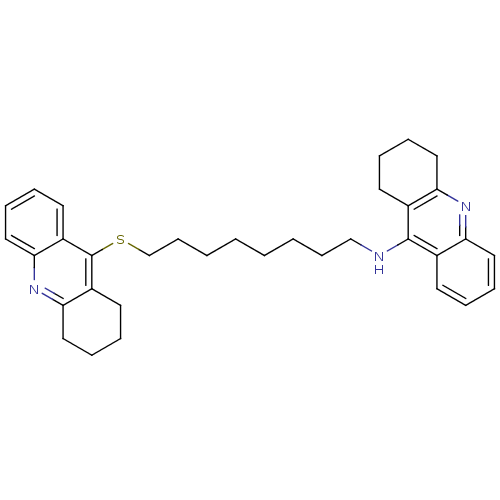
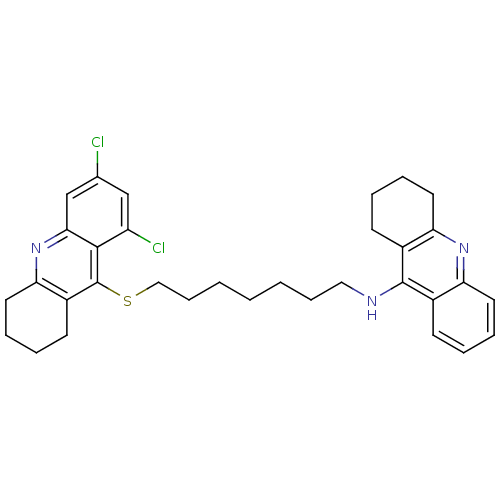
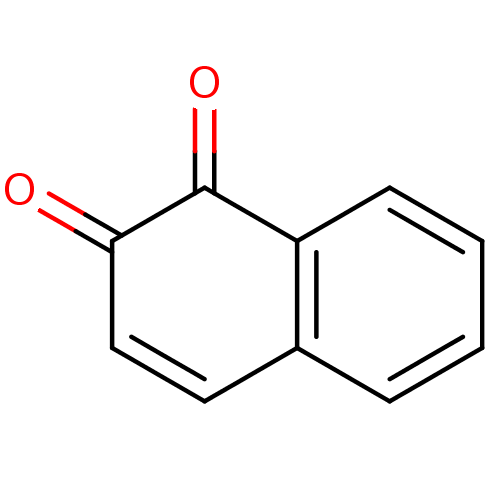
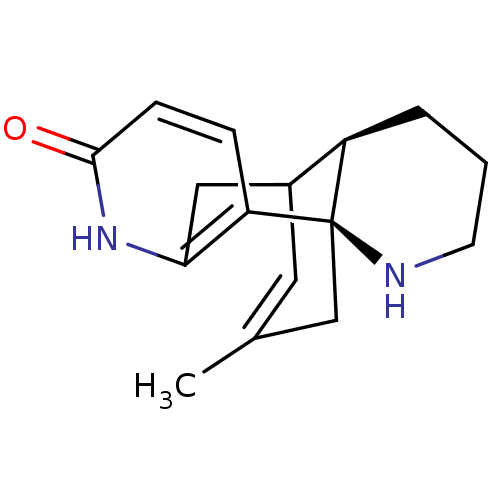
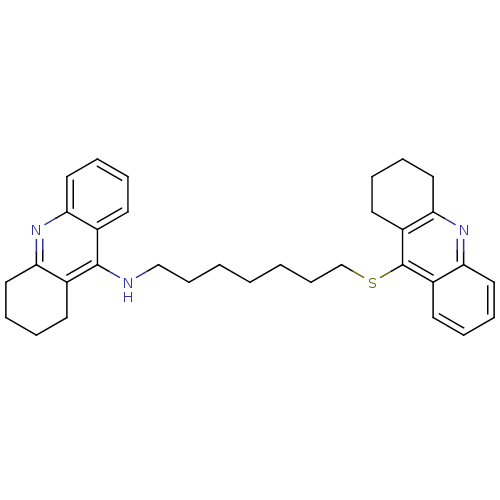
Affinity DataKi: 0.0260nM ΔG°: -14.4kcal/molepH: 8.0 T: 2°CAssay Description:The cholinesterase assays were performed using colorimetric method reported by E llman. Inhibition of enzyme activity was measured over a substrate c...More data for this Ligand-Target Pair
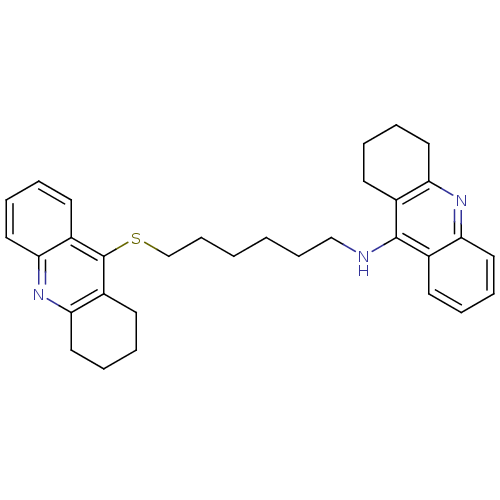
Affinity DataKi: 0.0600nM ΔG°: -13.9kcal/molepH: 8.0 T: 2°CAssay Description:Inhibition of enzyme activity was measured over a substrate concentration range of 0.01-30 mM and at least six inhibitor concentrations to determine ...More data for this Ligand-Target Pair
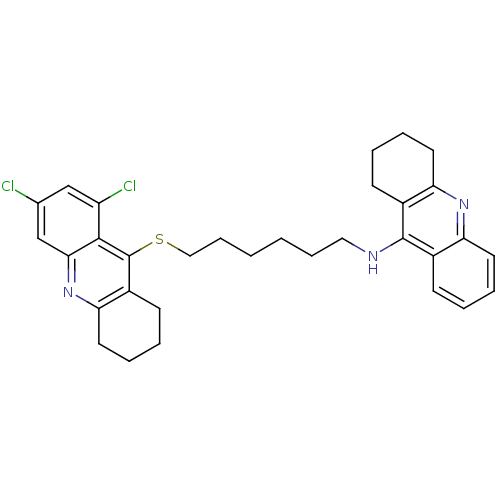
Affinity DataKi: 0.650nM ΔG°: -12.5kcal/molepH: 8.0 T: 2°CAssay Description:Inhibition of enzyme activity was measured over a substrate concentration range of 0.01-30 mM and at least six inhibitor concentrations to determine ...More data for this Ligand-Target Pair
TargetAcetylcholinesterase(Tetronarce californica (Pacific electric ray) (Tor...)
Weizmann Institute Of Science
Weizmann Institute Of Science
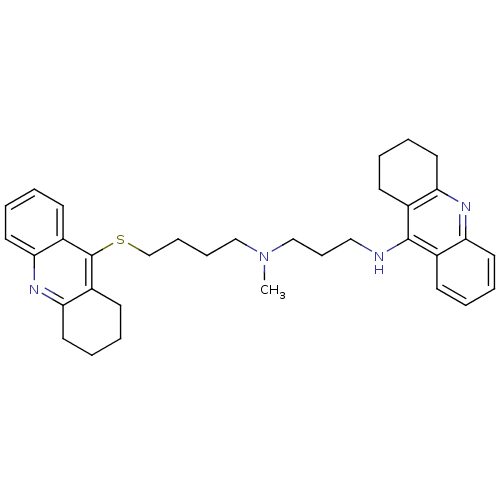
Affinity DataKi: 0.800nM ΔG°: -12.3kcal/mole IC50: 2.40nMpH: 7.0 T: 2°CAssay Description:The cholinesterase assays were performed using colorimetric method reported by Ellman. Enzyme activity was determined by measuring the absorbance at ...More data for this Ligand-Target Pair
Affinity DataKi: 1nM ΔG°: -12.3kcal/molepH: 8.0 T: 2°CAssay Description:Inhibition of enzyme activity was measured over a substrate concentration range of 0.01-30 mM and at least six inhibitor concentrations to determine ...More data for this Ligand-Target Pair
Affinity DataKi: 1.30nM ΔG°: -12.1kcal/molepH: 8.0 T: 2°CAssay Description:Inhibition of enzyme activity was measured over a substrate concentration range of 0.01-30 mM and at least six inhibitor concentrations to determine ...More data for this Ligand-Target Pair
Affinity DataKi: 1.30nM ΔG°: -12.1kcal/molepH: 8.0 T: 2°CAssay Description:Inhibition of enzyme activity was measured over a substrate concentration range of 0.01-30 mM and at least six inhibitor concentrations to determine ...More data for this Ligand-Target Pair
Affinity DataKi: 1.60nM ΔG°: -12.0kcal/molepH: 8.0 T: 2°CAssay Description:Inhibition of enzyme activity was measured over a substrate concentration range of 0.01-30 mM and at least six inhibitor concentrations to determine ...More data for this Ligand-Target Pair
Affinity DataKi: 1.90nM ΔG°: -11.9kcal/molepH: 8.0 T: 2°CAssay Description:Inhibition of enzyme activity was measured over a substrate concentration range of 0.01-30 mM and at least six inhibitor concentrations to determine ...More data for this Ligand-Target Pair
Affinity DataKi: 2.80nM ΔG°: -11.7kcal/molepH: 8.0 T: 2°CAssay Description:Inhibition of enzyme activity was measured over a substrate concentration range of 0.01-30 mM and at least six inhibitor concentrations to determine ...More data for this Ligand-Target Pair
Affinity DataKi: 2.90nM ΔG°: -11.6kcal/molepH: 8.0 T: 2°CAssay Description:Inhibition of enzyme activity was measured over a substrate concentration range of 0.01-30 mM and at least six inhibitor concentrations to determine ...More data for this Ligand-Target Pair
TargetAcetylcholinesterase(Electrophorus electricus (Electric eel))
Rheinische Friedrich-Wilhelms-Universitat Bonn
Rheinische Friedrich-Wilhelms-Universitat Bonn
Affinity DataKi: 3.23nM IC50: 3.25nMAssay Description:The cholinesterase assays were performed using colorimetric method reported by Ellman. The absorbance changed at 412 nm and was monitored with spectr...More data for this Ligand-Target Pair
TargetAcetylcholinesterase(Tetronarce californica (Pacific electric ray) (Tor...)
Weizmann Institute Of Science
Weizmann Institute Of Science
Affinity DataKi: 4.5nM ΔG°: -11.3kcal/mole IC50: 16nMpH: 7.0 T: 2°CAssay Description:The cholinesterase assays were performed using colorimetric method reported by Ellman. Enzyme activity was determined by measuring the absorbance at ...More data for this Ligand-Target Pair
Affinity DataKi: 6nM ΔG°: -11.2kcal/molepH: 8.0 T: 2°CAssay Description:Inhibition of enzyme activity was measured over a substrate concentration range of 0.01-30 mM and at least six inhibitor concentrations to determine ...More data for this Ligand-Target Pair
Affinity DataKi: 6.40nM ΔG°: -11.2kcal/molepH: 8.0 T: 2°CAssay Description:The cholinesterase assays were performed using colorimetric method reported by E llman. Inhibition of enzyme activity was measured over a substrate c...More data for this Ligand-Target Pair
Affinity DataKi: 6.90nM ΔG°: -11.0kcal/molepH: 7.4 T: 2°CAssay Description:The cholinesterase assays were performed using colorimetric method reported by Ellman. Estimates of the competitive inhibition constants (Ki) were ob...More data for this Ligand-Target Pair
Affinity DataKi: 9.10nM ΔG°: -11.0kcal/molepH: 8.0 T: 2°CAssay Description:Inhibition of enzyme activity was measured over a substrate concentration range of 0.01-30 mM and at least six inhibitor concentrations to determine ...More data for this Ligand-Target Pair
Affinity DataKi: 15.7nM ΔG°: -10.6kcal/molepH: 8.0 T: 2°CAssay Description:The cholinesterase assays were performed using colorimetric method reported by E llman. Inhibition of enzyme activity was measured over a substrate c...More data for this Ligand-Target Pair
Affinity DataKi: 16.5nM ΔG°: -10.6kcal/molepH: 8.0 T: 2°CAssay Description:The cholinesterase assays were performed using colorimetric method reported by E llman. Inhibition of enzyme activity was measured over a substrate c...More data for this Ligand-Target Pair
Affinity DataKi: 19.6nM ΔG°: -10.4kcal/mole IC50: 52nMpH: 7.0 T: 2°CAssay Description:The cholinesterase assays were performed using colorimetric method reported by Ellman. Enzyme activity was determined by measuring the absorbance at ...More data for this Ligand-Target Pair
Affinity DataKi: 20.5nM ΔG°: -10.9kcal/mole IC50: 23.1nMpH: 8.0 T: 2°CAssay Description:The cholinesterase assays were performed using colorimetric method reported by Ellman. Estimates of the competitive inhibition constants (Ki) were ob...More data for this Ligand-Target Pair
Affinity DataKi: 21.7nM ΔG°: -10.9kcal/mole IC50: 44.5nMpH: 8.0 T: 2°CAssay Description:The cholinesterase assays were performed using colorimetric method reported by Ellman. Estimates of the competitive inhibition constants (Ki) were ob...More data for this Ligand-Target Pair
Affinity DataKi: 30nM ΔG°: -10.3kcal/molepH: 8.0 T: 2°CAssay Description:Inhibition of enzyme activity was measured over a substrate concentration range of 0.01-30 mM and at least six inhibitor concentrations to determine ...More data for this Ligand-Target Pair
Affinity DataKi: 40nM ΔG°: -10.1kcal/molepH: 8.0 T: 2°CAssay Description:Inhibition of enzyme activity was measured over a substrate concentration range of 0.01-30 mM and at least six inhibitor concentrations to determine ...More data for this Ligand-Target Pair
Affinity DataKi: 40nM ΔG°: -10.1kcal/molepH: 8.0 T: 2°CAssay Description:Inhibition of enzyme activity was measured over a substrate concentration range of 0.01-30 mM and at least six inhibitor concentrations to determine ...More data for this Ligand-Target Pair
Affinity DataKi: 41nM ΔG°: -10.1kcal/molepH: 8.0 T: 2°CAssay Description:Inhibition of enzyme activity was measured over a substrate concentration range of 0.01-30 mM and at least six inhibitor concentrations to determine ...More data for this Ligand-Target Pair
Affinity DataKi: 47nM ΔG°: -9.99kcal/molepH: 8.0 T: 2°CAssay Description:Inhibition of enzyme activity was measured over a substrate concentration range of 0.01-30 mM and at least six inhibitor concentrations to determine ...More data for this Ligand-Target Pair
Affinity DataKi: 47nM ΔG°: -9.99kcal/molepH: 8.0 T: 2°CAssay Description:The cholinesterase assays were performed using colorimetric method reported by E llman. Inhibition of enzyme activity was measured over a substrate c...More data for this Ligand-Target Pair
Affinity DataKi: 47.1nM ΔG°: -9.89kcal/mole IC50: 114nMpH: 7.0 T: 2°CAssay Description:The cholinesterase assays were performed using colorimetric method reported by Ellman. Enzyme activity was determined by measuring the absorbance at ...More data for this Ligand-Target Pair
Affinity DataKi: 49nM ΔG°: -9.86kcal/molepH: 7.4 T: 2°CAssay Description:The cholinesterase assays were performed using colorimetric method reported by Ellman. Estimates of the competitive inhibition constants (Ki) were ob...More data for this Ligand-Target Pair
Affinity DataKi: 50nM ΔG°: -9.95kcal/molepH: 8.0 T: 2°CAssay Description:Inhibition of enzyme activity was measured over a substrate concentration range of 0.01-30 mM and at least six inhibitor concentrations to determine ...More data for this Ligand-Target Pair
Affinity DataKi: 100nM ΔG°: -9.54kcal/molepH: 8.0 T: 2°CAssay Description:Inhibition of enzyme activity was measured over a substrate concentration range of 0.01-30 mM and at least six inhibitor concentrations to determine ...More data for this Ligand-Target Pair
Affinity DataKi: 120nM ΔG°: -9.43kcal/molepH: 8.0 T: 2°CAssay Description:Inhibition of enzyme activity was measured over a substrate concentration range of 0.01-30 mM and at least six inhibitor concentrations to determine ...More data for this Ligand-Target Pair
Affinity DataKi: 130nM ΔG°: -9.39kcal/molepH: 8.0 T: 2°CAssay Description:Inhibition of enzyme activity was measured over a substrate concentration range of 0.01-30 mM and at least six inhibitor concentrations to determine ...More data for this Ligand-Target Pair
Affinity DataKi: 137nM ΔG°: -9.36kcal/molepH: 8.0 T: 2°CAssay Description:The cholinesterase assays were performed using colorimetric method reported by E llman. Inhibition of enzyme activity was measured over a substrate c...More data for this Ligand-Target Pair
Affinity DataKi: 150nM ΔG°: -9.30kcal/molepH: 8.0 T: 2°CAssay Description:Inhibition of enzyme activity was measured over a substrate concentration range of 0.01-30 mM and at least six inhibitor concentrations to determine ...More data for this Ligand-Target Pair
Affinity DataKi: 151nM IC50: 250nMAssay Description:The cholinesterase assays were performed using colorimetric method reported by Ellman. The absorbance changes at 412 nm were recorded for 5 min with ...More data for this Ligand-Target Pair
Affinity DataKi: 155nM ΔG°: -9.66kcal/mole IC50: 0.253nMpH: 8.0 T: 2°CAssay Description:The cholinesterase assays were performed using colorimetric method reported by Ellman. Enzyme activity was determined by measuring the absorbance at...More data for this Ligand-Target Pair
Affinity DataKi: 163nMAssay Description:The cholinesterase assays were performed using colorimetric method reported by Ellman. Estimates of the competitive inhibition constants (Ki) were ob...More data for this Ligand-Target Pair
TargetAcetylcholinesterase(Tetronarce californica (Pacific electric ray) (Tor...)
Weizmann Institute Of Science
Weizmann Institute Of Science
Affinity DataKi: 175nM ΔG°: -9.12kcal/mole IC50: 414nMpH: 7.0 T: 2°CAssay Description:The cholinesterase assays were performed using colorimetric method reported by Ellman. Enzyme activity was determined by measuring the absorbance at ...More data for this Ligand-Target Pair
TargetAcetylcholinesterase(Tetronarce californica (Pacific electric ray) (Tor...)
Weizmann Institute Of Science
Weizmann Institute Of Science
Affinity DataKi: 175nM ΔG°: -9.21kcal/molepH: 7.0 T: 2°CAssay Description:The cholinesterase assays were performed using colorimetric method reported by Ellman. Enzyme activity was determined by measuring the absorbance at ...More data for this Ligand-Target Pair
Affinity DataKi: 195nM ΔG°: -9.15kcal/molepH: 8.0 T: 2°CAssay Description:Inhibition of enzyme activity was measured over a substrate concentration range of 0.01-30 mM and at least six inhibitor concentrations to determine ...More data for this Ligand-Target Pair
Affinity DataKi: 210nM IC50: 680nMAssay Description:The cholinesterase assays were performed using colorimetric method reported by Ellman. The absorbance changes at 412 nm were recorded for 5 min with ...More data for this Ligand-Target Pair
Affinity DataKi: 210nM ΔG°: -9.10kcal/molepH: 8.0 T: 2°CAssay Description:Inhibition of enzyme activity was measured over a substrate concentration range of 0.01-30 mM and at least six inhibitor concentrations to determine ...More data for this Ligand-Target Pair
Affinity DataKi: 250nM ΔG°: -9.00kcal/molepH: 8.0 T: 2°CAssay Description:Inhibition of enzyme activity was measured over a substrate concentration range of 0.01-30 mM and at least six inhibitor concentrations to determine ...More data for this Ligand-Target Pair
Affinity DataKi: 290nM ΔG°: -8.91kcal/molepH: 8.0 T: 2°CAssay Description:Inhibition of enzyme activity was measured over a substrate concentration range of 0.01-30 mM and at least six inhibitor concentrations to determine ...More data for this Ligand-Target Pair
Affinity DataKi: 320nMAssay Description:The cholinesterase assays were performed using colorimetric method reported by Ellman. Estimates of the competitive inhibition constants (Ki) were ob...More data for this Ligand-Target Pair
TargetAcetylcholinesterase(Tetronarce californica (Pacific electric ray) (Tor...)
Weizmann Institute Of Science
Weizmann Institute Of Science
Affinity DataKi: 334nM ΔG°: -8.83kcal/molepH: 7.0 T: 2°CAssay Description:The cholinesterase assays were performed using colorimetric method reported by Ellman. Enzyme activity was determined by measuring the absorbance at ...More data for this Ligand-Target Pair
Affinity DataKi: 340nM ΔG°: -8.82kcal/molepH: 8.0 T: 2°CAssay Description:Inhibition of enzyme activity was measured over a substrate concentration range of 0.01-30 mM and at least six inhibitor concentrations to determine ...More data for this Ligand-Target Pair
Affinity DataKi: 340nM ΔG°: -8.82kcal/molepH: 8.0 T: 2°CAssay Description:Inhibition of enzyme activity was measured over a substrate concentration range of 0.01-30 mM and at least six inhibitor concentrations to determine ...More data for this Ligand-Target Pair

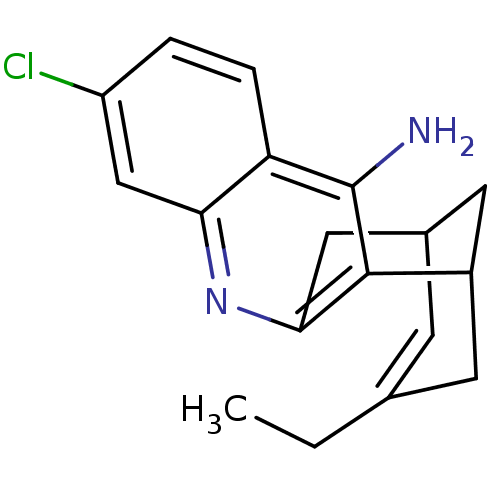
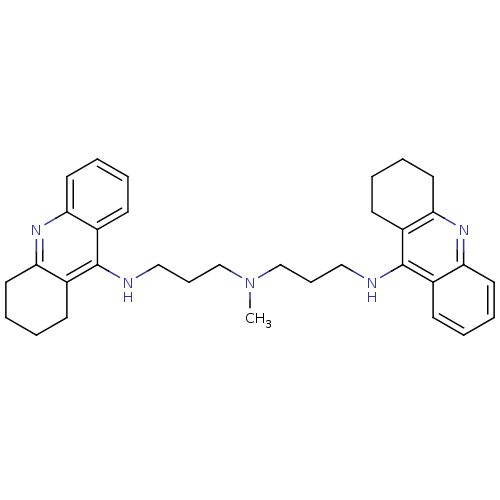
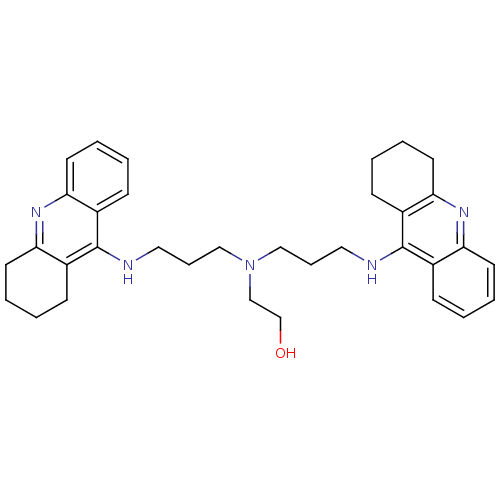
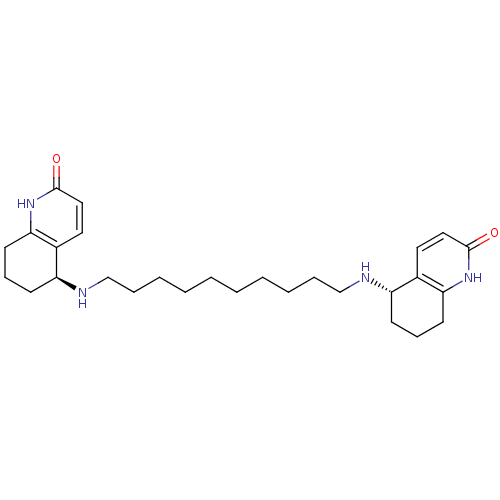
 3D Structure (crystal)
3D Structure (crystal)