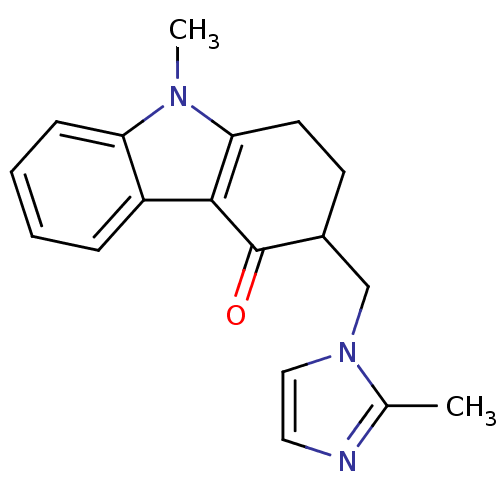
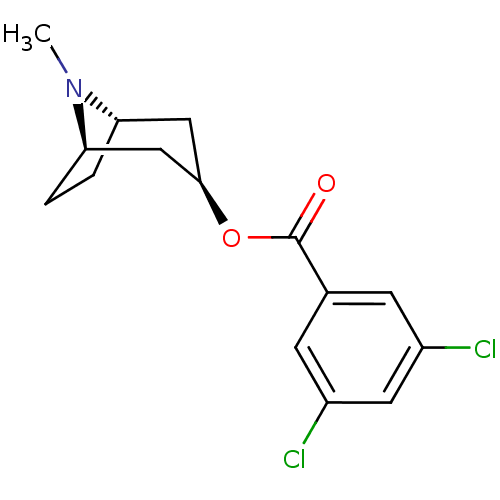
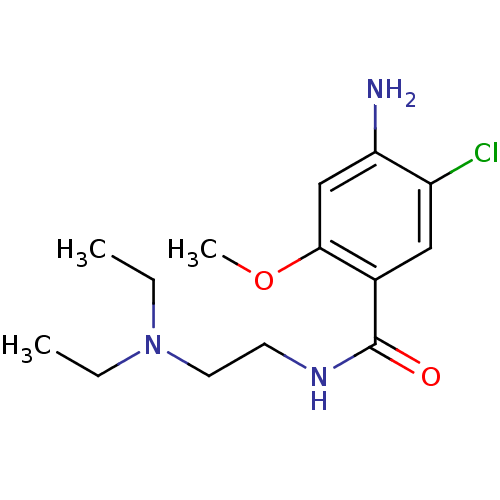
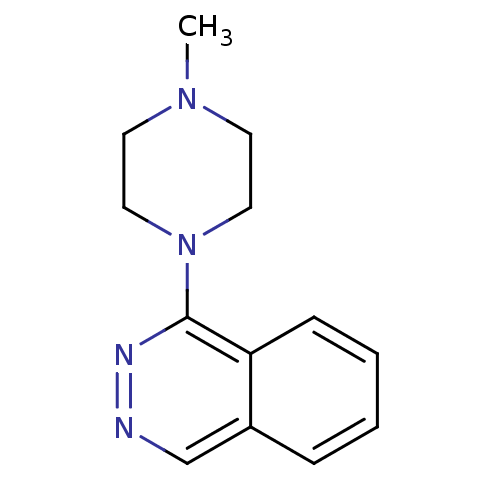
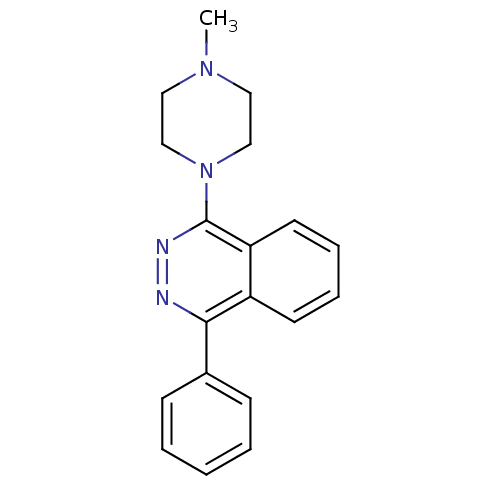
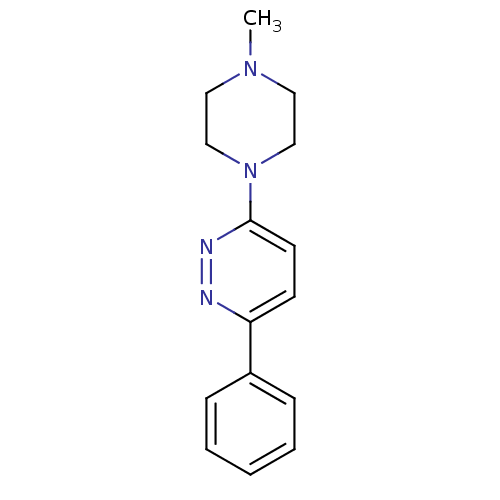
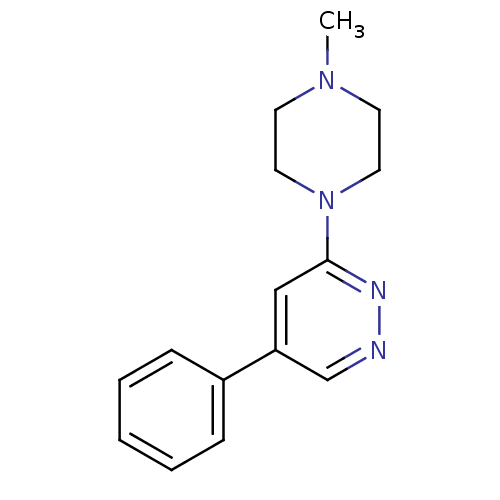
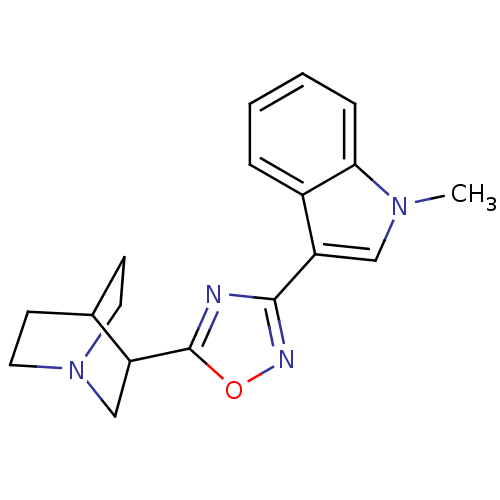
Affinity DataKi: 0.600nMAssay Description:Compound was evaluated for its binding affinity for 5-hydroxytryptamine 3 receptor by measuring displacement [3H]GR-65630 in rat cerebral cortexMore data for this Ligand-Target Pair
Affinity DataKi: 1.40nMAssay Description:Compound was evaluated for its binding affinity for 5-hydroxytryptamine 3 receptor by measuring displacement [3H]GR-65630 in rat cerebral cortexMore data for this Ligand-Target Pair
Affinity DataKi: 3.20nMAssay Description:Compound was evaluated for its binding affinity for 5-hydroxytryptamine 3 receptor by measuring displacement [3H]GR-65630 in rat cerebral cortexMore data for this Ligand-Target Pair
Affinity DataKi: 3.30nMAssay Description:Compound was evaluated for its binding affinity for 5-hydroxytryptamine 3 receptor by measuring displacement [3H]GR-65630 in rat cerebral cortexMore data for this Ligand-Target Pair
Affinity DataKi: 54nMAssay Description:Compound was evaluated for its binding affinity for 5-hydroxytryptamine 3 receptor by measuring displacement [3H]GR-65630 in rat cerebral cortexMore data for this Ligand-Target Pair
Affinity DataKi: 348nMAssay Description:Compound was evaluated for its binding affinity for 5-hydroxytryptamine 3 receptor by measuring displacement [3H]GR-65630 in rat cerebral cortexMore data for this Ligand-Target Pair
Affinity DataKi: 1.00E+4nMAssay Description:Compound was evaluated for its binding affinity for Muscarinic acetylcholine receptor M1 by measuring displacement of [3H]- pirenzepine from rat hipp...More data for this Ligand-Target Pair
Affinity DataKi: 1.30E+4nMAssay Description:Compound was evaluated for its binding affinity for Muscarinic acetylcholine receptor M1 by measuring displacement of [3H]- pirenzepine from rat hipp...More data for this Ligand-Target Pair
Affinity DataKi: 1.56E+4nMAssay Description:Compound was evaluated for its binding affinity for Muscarinic acetylcholine receptor M1 by measuring displacement of [3H]- pirenzepine from rat hipp...More data for this Ligand-Target Pair
Affinity DataKi: 3.80E+4nMAssay Description:Compound was evaluated for its binding affinity for Muscarinic acetylcholine receptor M2 by measuring displacement of [3H]- NMS ligand from rat cardi...More data for this Ligand-Target Pair
Affinity DataKi: 6.20E+4nMAssay Description:Compound was evaluated for its binding affinity for Muscarinic acetylcholine receptor M1 by measuring displacement of [3H]- pirenzepine from rat hipp...More data for this Ligand-Target Pair
Affinity DataKi: 7.10E+4nMAssay Description:Compound was evaluated for its binding affinity for Muscarinic acetylcholine receptor M2 by measuring displacement of [3H]- NMS ligand from rat cardi...More data for this Ligand-Target Pair
Affinity DataKi: 8.00E+4nMAssay Description:Compound was evaluated for its binding affinity for Muscarinic acetylcholine receptor M2 by measuring displacement of [3H]- NMS ligand from rat cardi...More data for this Ligand-Target Pair
Affinity DataKi: 1.16E+5nMAssay Description:Compound was evaluated for its binding affinity for Muscarinic acetylcholine receptor M2 by measuring displacement of [3H]- NMS ligand from rat cardi...More data for this Ligand-Target Pair
Affinity DataIC50: 1.40nMAssay Description:Compound was evaluated for its binding affinity for 5-hydroxytryptamine 3 receptor by measuring displacement [3H]GR-65630 in rat cerebral cortex.More data for this Ligand-Target Pair
Affinity DataIC50: 10nMAssay Description:Compound was evaluated for its binding affinity for 5-hydroxytryptamine 3 receptor by measuring displacement of [3H]granisetron from rat cerebral cor...More data for this Ligand-Target Pair
Affinity DataIC50: 36nMAssay Description:Compound was evaluated for its binding affinity for 5-hydroxytryptamine 3 receptor by measuring displacement of [3H]granisetron from rat cerebral cor...More data for this Ligand-Target Pair
Affinity DataIC50: 370nMAssay Description:Compound was evaluated for its binding affinity for 5-hydroxytryptamine 3 receptor by measuring displacement of [3H]granisetron from rat cerebral cor...More data for this Ligand-Target Pair
Affinity DataIC50: 425nMAssay Description:Compound was evaluated for its binding affinity for 5-hydroxytryptamine 3 receptor by measuring displacement of [3H]granisetron from rat cerebral cor...More data for this Ligand-Target Pair

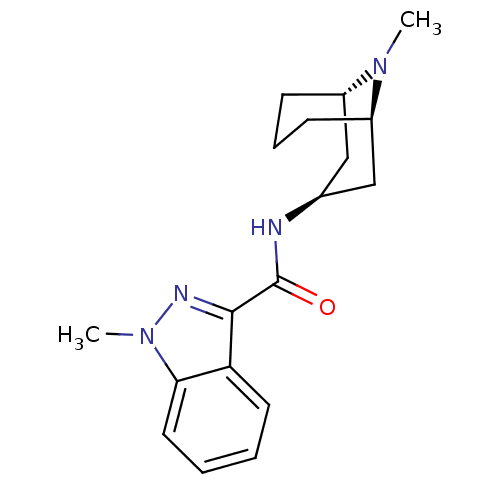
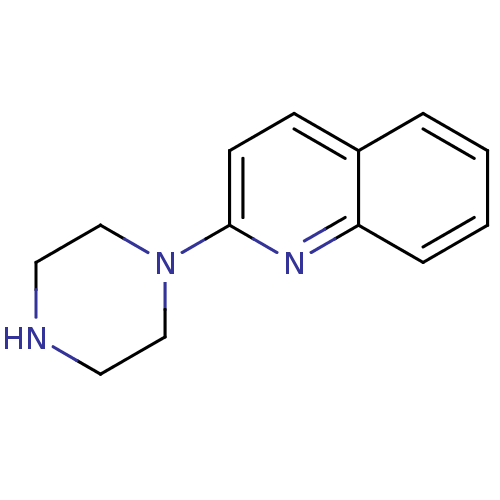
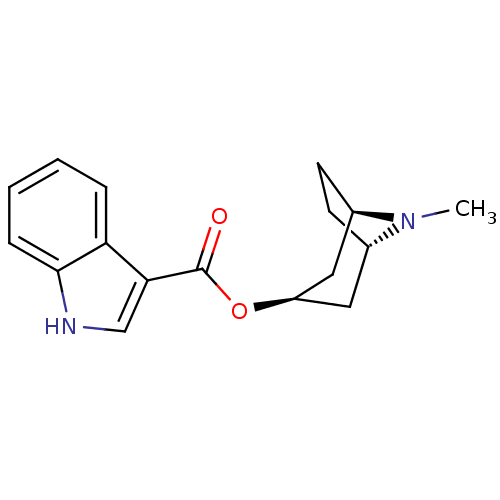
 3D Structure (crystal)
3D Structure (crystal)