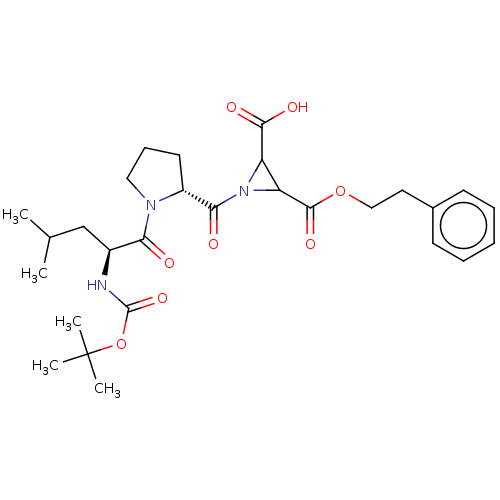
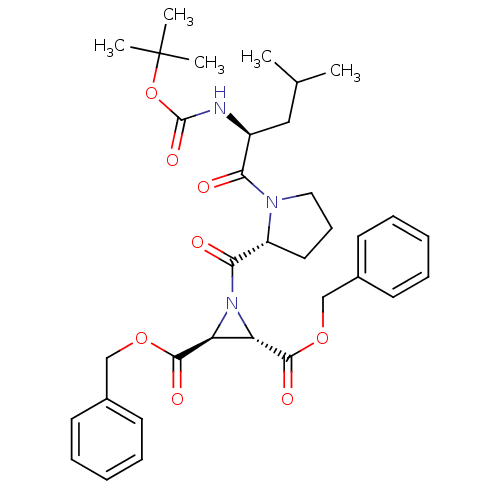
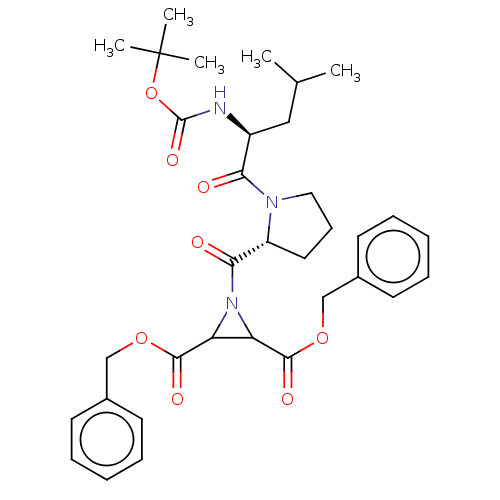
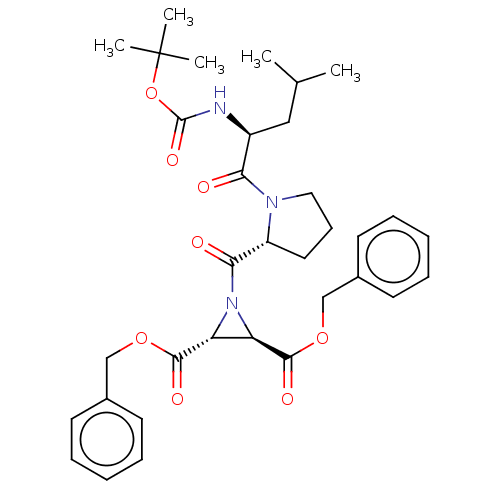
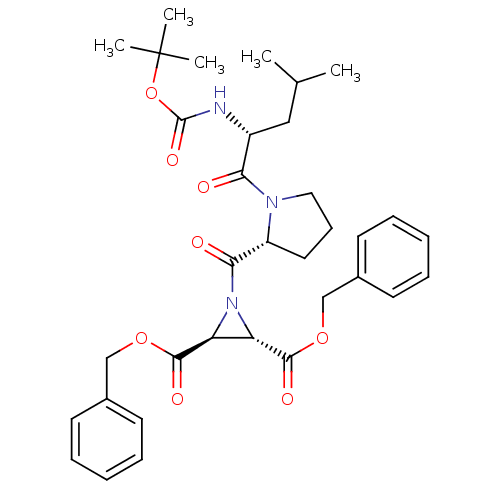
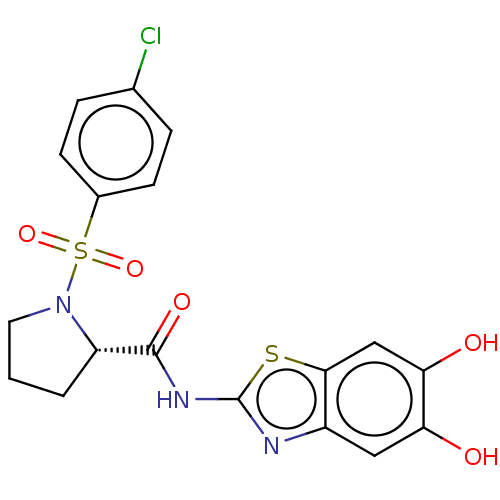
TargetCysteine protease(Trypanosoma brucei rhodesiense)
Johannes Gutenberg-Universit£T Mainz
Curated by ChEMBL
Johannes Gutenberg-Universit£T Mainz
Curated by ChEMBL
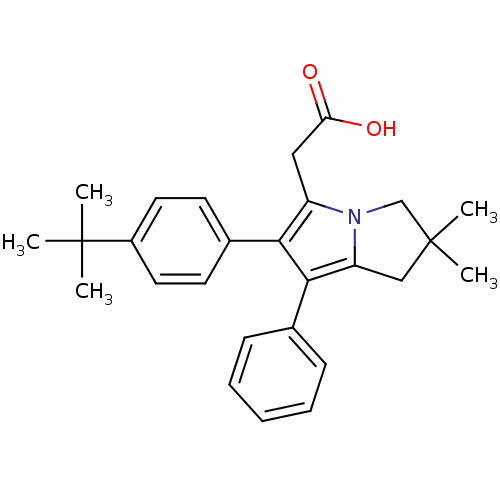
Affinity DataKi: 120nMAssay Description:Inhibition of Trypanosoma brucei rhodesiense rhodesain using Z-Phe-Arg-AMC as substrate by fluorometric assayMore data for this Ligand-Target Pair
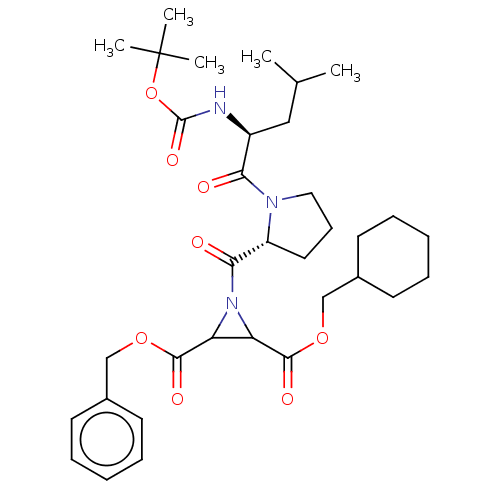
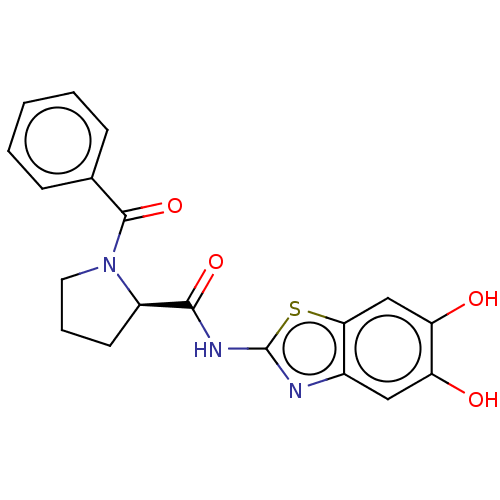
TargetCysteine protease(Trypanosoma brucei rhodesiense)
Johannes Gutenberg-Universit£T Mainz
Curated by ChEMBL
Johannes Gutenberg-Universit£T Mainz
Curated by ChEMBL
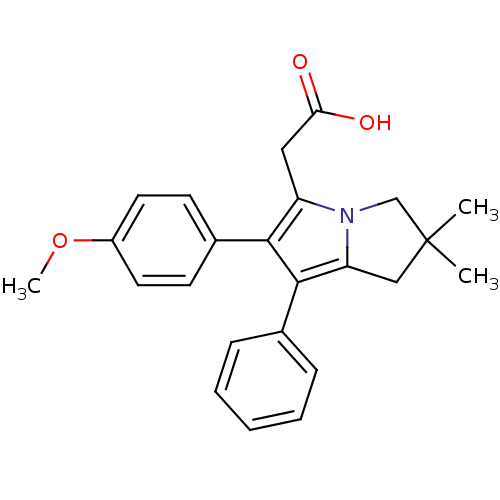
Affinity DataKi: 160nMAssay Description:Inhibition of Trypanosoma brucei rhodesiense rhodesain using Z-Phe-Arg-AMC as substrate by fluorometric assayMore data for this Ligand-Target Pair
Affinity DataKi: 400nMAssay Description:Inhibition of Plasmodium falciparum falcipain 2 using Z-Phe-Arg-AMC as substrate by fluorometric assayMore data for this Ligand-Target Pair
Affinity DataKi: 400nMAssay Description:Inhibition of human cathepsin-L using Z-Phe-Arg-AMC as substrate by fluorometric assayMore data for this Ligand-Target Pair
Affinity DataKi: 450nMAssay Description:Inhibition of Plasmodium falciparum falcipain 2 using Z-Phe-Arg-AMC as substrate by fluorometric assayMore data for this Ligand-Target Pair
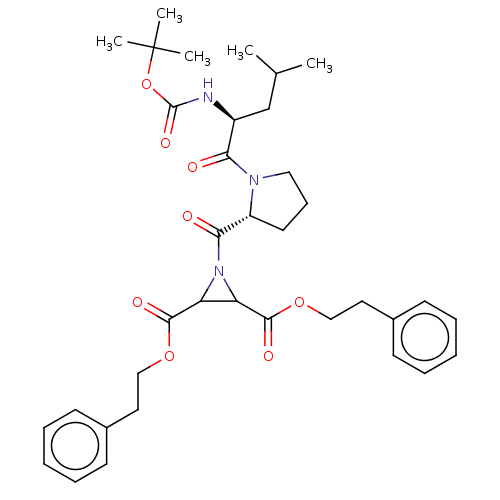
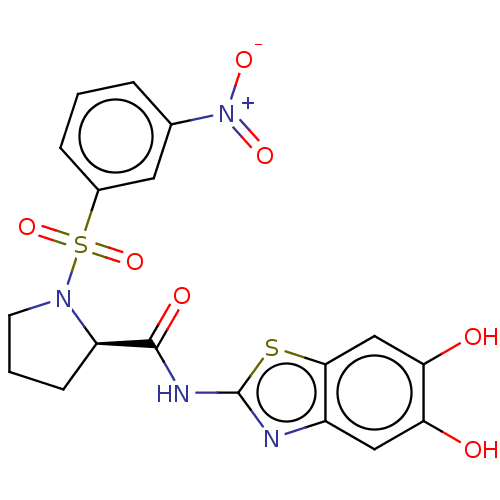
TargetCysteine protease(Trypanosoma brucei rhodesiense)
Johannes Gutenberg-Universit£T Mainz
Curated by ChEMBL
Johannes Gutenberg-Universit£T Mainz
Curated by ChEMBL
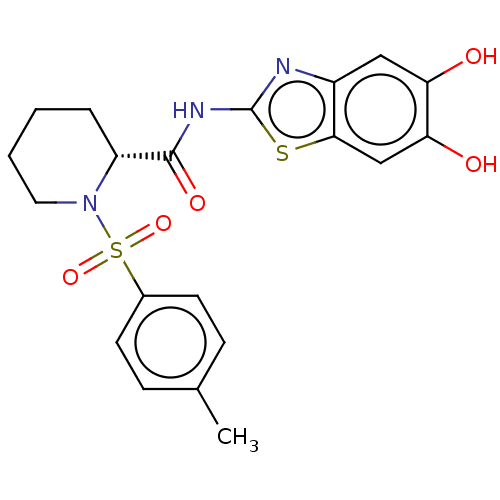
Affinity DataKi: 500nMAssay Description:Inhibition of Trypanosoma brucei rhodesiense rhodesain using Z-Phe-Arg-AMC as substrate by fluorometric assayMore data for this Ligand-Target Pair
Affinity DataKi: 710nMAssay Description:Inhibition of Trypanosoma cruzi cruzain using Z-Phe-Arg-AMC as substrate by fluorometric assayMore data for this Ligand-Target Pair
Affinity DataKi: 790nMAssay Description:Inhibition of Trypanosoma cruzi cruzain using Z-Phe-Arg-AMC as substrate by fluorometric assayMore data for this Ligand-Target Pair
Affinity DataKi: 900nMAssay Description:Inhibition of Trypanosoma cruzi cruzain using Z-Phe-Arg-AMC as substrate by fluorometric assayMore data for this Ligand-Target Pair
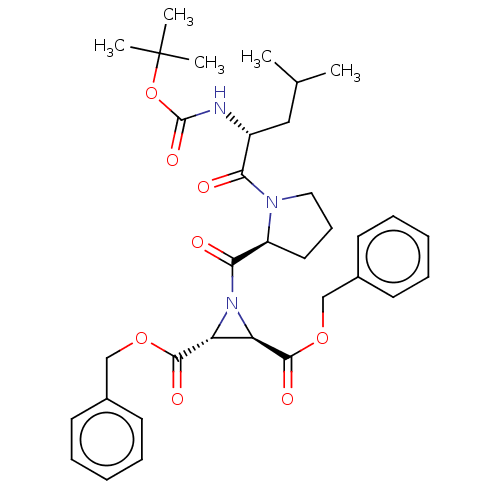
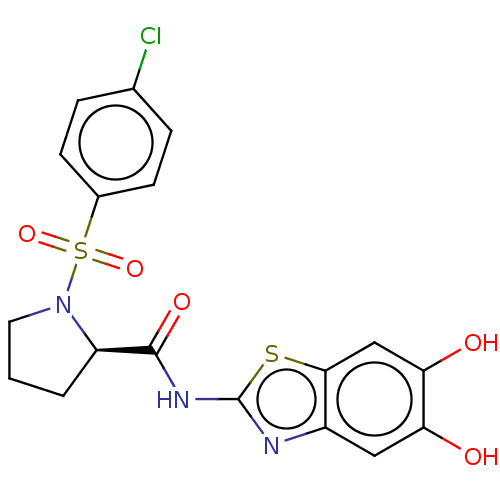
TargetCysteine protease(Trypanosoma brucei rhodesiense)
Johannes Gutenberg-Universit£T Mainz
Curated by ChEMBL
Johannes Gutenberg-Universit£T Mainz
Curated by ChEMBL
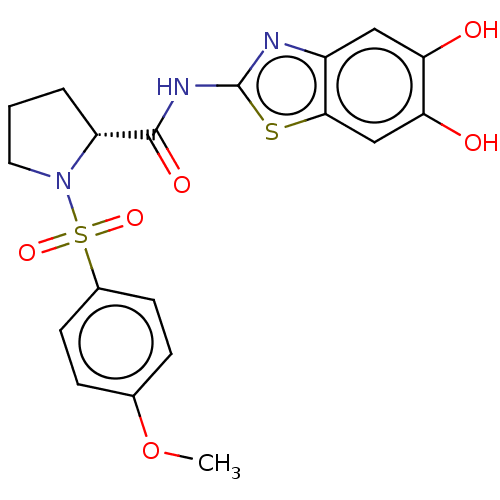
Affinity DataKi: 1.10E+3nMAssay Description:Inhibition of Trypanosoma brucei rhodesiense rhodesain using Z-Phe-Arg-AMC as substrate by fluorometric assayMore data for this Ligand-Target Pair
TargetCysteine protease(Trypanosoma brucei rhodesiense)
Johannes Gutenberg-Universit£T Mainz
Curated by ChEMBL
Johannes Gutenberg-Universit£T Mainz
Curated by ChEMBL
Affinity DataKi: 1.20E+3nMAssay Description:Inhibition of Trypanosoma brucei rhodesiense rhodesain using Z-Phe-Arg-AMC as substrate by fluorometric assayMore data for this Ligand-Target Pair
Affinity DataKi: 1.26E+3nMAssay Description:Inhibition of Plasmodium falciparum falcipain 2 using Z-Phe-Arg-AMC as substrate by fluorometric assayMore data for this Ligand-Target Pair
TargetCysteine protease(Trypanosoma brucei rhodesiense)
Johannes Gutenberg-Universit£T Mainz
Curated by ChEMBL
Johannes Gutenberg-Universit£T Mainz
Curated by ChEMBL
Affinity DataKi: 1.50E+3nMAssay Description:Inhibition of Trypanosoma brucei rhodesiense rhodesain using Z-Phe-Arg-AMC as substrate by fluorometric assayMore data for this Ligand-Target Pair
TargetCysteine protease(Trypanosoma brucei rhodesiense)
Johannes Gutenberg-Universit£T Mainz
Curated by ChEMBL
Johannes Gutenberg-Universit£T Mainz
Curated by ChEMBL
Affinity DataKi: 1.70E+3nMAssay Description:Inhibition of Trypanosoma brucei rhodesiense rhodesain using Z-Phe-Arg-AMC as substrate by fluorometric assayMore data for this Ligand-Target Pair
Affinity DataKi: 1.80E+3nMAssay Description:Inhibition of Trypanosoma cruzi cruzain using Z-Phe-Arg-AMC as substrate by fluorometric assayMore data for this Ligand-Target Pair
TargetCysteine protease(Trypanosoma brucei rhodesiense)
Johannes Gutenberg-Universit£T Mainz
Curated by ChEMBL
Johannes Gutenberg-Universit£T Mainz
Curated by ChEMBL
Affinity DataKi: 2.30E+3nMAssay Description:Inhibition of Trypanosoma brucei rhodesiense rhodesain using Z-Phe-Arg-AMC as substrate by fluorometric assayMore data for this Ligand-Target Pair
TargetCysteine protease(Trypanosoma brucei rhodesiense)
Johannes Gutenberg-Universit£T Mainz
Curated by ChEMBL
Johannes Gutenberg-Universit£T Mainz
Curated by ChEMBL
Affinity DataKi: 2.80E+3nMAssay Description:Inhibition of Trypanosoma brucei rhodesiense rhodesain using Z-Phe-Arg-AMC as substrate by fluorometric assayMore data for this Ligand-Target Pair
Affinity DataKi: 4.00E+3nMAssay Description:Inhibition of human cathepsin-L using Z-Phe-Arg-AMC as substrate by fluorometric assayMore data for this Ligand-Target Pair
Affinity DataKi: 5.50E+3nMAssay Description:Inhibition of Trypanosoma cruzi cruzain using Z-Phe-Arg-AMC as substrate by fluorometric assayMore data for this Ligand-Target Pair
Affinity DataKi: 6.00E+3nMAssay Description:Inhibition of human cathepsin-L using Z-Phe-Arg-AMC as substrate by fluorometric assayMore data for this Ligand-Target Pair
Affinity DataKi: 7.30E+3nMAssay Description:Inhibition of human cathepsin-L using Z-Phe-Arg-AMC as substrate by fluorometric assayMore data for this Ligand-Target Pair
Affinity DataKi: 7.60E+3nMAssay Description:Inhibition of Trypanosoma cruzi cruzain using Z-Phe-Arg-AMC as substrate by fluorometric assayMore data for this Ligand-Target Pair
Affinity DataKi: 7.70E+3nMAssay Description:Inhibition of Trypanosoma cruzi cruzain using Z-Phe-Arg-AMC as substrate by fluorometric assayMore data for this Ligand-Target Pair
Affinity DataKi: 1.20E+4nMAssay Description:Inhibition of human cathepsin-L using Z-Phe-Arg-AMC as substrate by fluorometric assayMore data for this Ligand-Target Pair
TargetCysteine protease(Trypanosoma brucei rhodesiense)
Johannes Gutenberg-Universit£T Mainz
Curated by ChEMBL
Johannes Gutenberg-Universit£T Mainz
Curated by ChEMBL
Affinity DataKi: 1.52E+4nMAssay Description:Inhibition of Trypanosoma brucei rhodesiense rhodesain using Z-Phe-Arg-AMC as substrate by fluorometric assayMore data for this Ligand-Target Pair
TargetCathepsin B-like protease(Leishmania major)
Johannes Gutenberg-Universit£T Mainz
Curated by ChEMBL
Johannes Gutenberg-Universit£T Mainz
Curated by ChEMBL
Affinity DataKi: 1.82E+4nMAssay Description:Inhibition of Leishmania major MHOM/IL/81/FE/BNI His6-tagged CPC expressed in Pichia pastoris using Cbz-Phe-Arg-AMC as substrate by fluorescence spec...More data for this Ligand-Target Pair
Affinity DataKi: 2.83E+4nMAssay Description:Inhibition of Plasmodium falciparum falcipain 2 using Z-Phe-Arg-AMC as substrate by fluorometric assayMore data for this Ligand-Target Pair
Affinity DataKi: 2.86E+4nMAssay Description:Inhibition of Plasmodium falciparum falcipain 2 using Z-Phe-Arg-AMC as substrate by fluorometric assayMore data for this Ligand-Target Pair
Affinity DataKi: 3.09E+4nMAssay Description:Inhibition of Trypanosoma cruzi cruzain using Z-Phe-Arg-AMC as substrate by fluorometric assayMore data for this Ligand-Target Pair
Affinity DataKi: 4.39E+4nMAssay Description:Inhibition of Plasmodium falciparum falcipain 2 using Z-Phe-Arg-AMC as substrate by fluorometric assayMore data for this Ligand-Target Pair
Affinity DataKi: 1.14E+5nMAssay Description:Inhibition of human cathepsin-B using Z-Phe-Arg-AMC as substrate by fluorometric assayMore data for this Ligand-Target Pair
Affinity DataIC50: 180nMAssay Description:Inhibition of 5-lipoxygenase in bovine PMNL cell assay.More data for this Ligand-Target Pair
Affinity DataIC50: 180nMAssay Description:Inhibition of 5-lipoxygenase in bovine PMNL cell assay.More data for this Ligand-Target Pair
Affinity DataIC50: 230nMAssay Description:Inhibition of human 5-lipoxygenase in human cellsMore data for this Ligand-Target Pair
Affinity DataIC50: 240nMAssay Description:Inhibition of 5-lipoxygenase in bovine PMNL cell assay.More data for this Ligand-Target Pair
Affinity DataIC50: 320nMAssay Description:Inhibition of N-terminal hexa-histidine tagged Zika virus NS2B/NS3 protease expressed in Escherichia coli BL21-Gold (DE3) using fluorogenic AMC-subst...More data for this Ligand-Target Pair
Affinity DataIC50: 510nMAssay Description:Inhibition of N-terminal hexa-histidine tagged Zika virus NS2B/NS3 protease expressed in Escherichia coli BL21-Gold (DE3) using fluorogenic AMC-subst...More data for this Ligand-Target Pair
Affinity DataIC50: 580nMAssay Description:Inhibition of N-terminal hexa-histidine tagged Zika virus NS2B/NS3 protease expressed in Escherichia coli BL21-Gold (DE3) using fluorogenic AMC-subst...More data for this Ligand-Target Pair
Affinity DataIC50: 810nMAssay Description:Inhibition of N-terminal hexa-histidine tagged Zika virus NS2B/NS3 protease expressed in Escherichia coli BL21-Gold (DE3) using fluorogenic AMC-subst...More data for this Ligand-Target Pair
Affinity DataIC50: 860nMAssay Description:Inhibition of N-terminal hexa-histidine tagged Zika virus NS2B/NS3 protease expressed in Escherichia coli BL21-Gold (DE3) using fluorogenic AMC-subst...More data for this Ligand-Target Pair
Affinity DataIC50: 930nMAssay Description:Inhibition of N-terminal hexa-histidine tagged Zika virus NS2B/NS3 protease expressed in Escherichia coli BL21-Gold (DE3) using fluorogenic AMC-subst...More data for this Ligand-Target Pair
Affinity DataIC50: 930nMAssay Description:Inhibition of N-terminal hexa-histidine tagged Zika virus NS2B/NS3 protease expressed in Escherichia coli BL21-Gold (DE3) using fluorogenic AMC-subst...More data for this Ligand-Target Pair
Affinity DataIC50: 940nMAssay Description:Inhibition of N-terminal hexa-histidine tagged Zika virus NS2B/NS3 protease expressed in Escherichia coli BL21-Gold (DE3) using fluorogenic AMC-subst...More data for this Ligand-Target Pair
Affinity DataIC50: 1.03E+3nMAssay Description:Inhibition of N-terminal hexa-histidine tagged Zika virus NS2B/NS3 protease expressed in Escherichia coli BL21-Gold (DE3) using fluorogenic AMC-subst...More data for this Ligand-Target Pair
Affinity DataIC50: 1.11E+3nMAssay Description:Inhibition of N-terminal hexa-histidine tagged Zika virus NS2B/NS3 protease expressed in Escherichia coli BL21-Gold (DE3) using fluorogenic AMC-subst...More data for this Ligand-Target Pair
Affinity DataIC50: 1.32E+3nMAssay Description:Inhibition of N-terminal hexa-histidine tagged Zika virus NS2B/NS3 protease expressed in Escherichia coli BL21-Gold (DE3) using fluorogenic AMC-subst...More data for this Ligand-Target Pair
Affinity DataIC50: 1.32E+3nMAssay Description:Inhibition of N-terminal hexa-histidine tagged Zika virus NS2B/NS3 protease expressed in Escherichia coli BL21-Gold (DE3) using fluorogenic AMC-subst...More data for this Ligand-Target Pair
Affinity DataIC50: 1.35E+3nMAssay Description:Inhibition of N-terminal hexa-histidine tagged Zika virus NS2B/NS3 protease expressed in Escherichia coli BL21-Gold (DE3) using fluorogenic AMC-subst...More data for this Ligand-Target Pair
Affinity DataIC50: 1.40E+3nMAssay Description:Inhibition of N-terminal hexa-histidine tagged Zika virus NS2B/NS3 protease expressed in Escherichia coli BL21-Gold (DE3) using fluorogenic AMC-subst...More data for this Ligand-Target Pair
Affinity DataIC50: 1.41E+3nMAssay Description:Inhibition of N-terminal hexa-histidine tagged Zika virus NS2B/NS3 protease expressed in Escherichia coli BL21-Gold (DE3) using fluorogenic AMC-subst...More data for this Ligand-Target Pair