Reaction Details Report a problem with these data
Report a problem with these data
 Report a problem with these data
Report a problem with these dataTarget
Alpha-glucosidase MAL32
Ligand
BDBM23838
Substrate
BDBM23837
Meas. Tech.
alpha-Glucosidase Inhibition Assay
pH
7±n/a
Temperature
310.15±n/a K
IC50
100000±n/a nM
Citation
 Dodo, K; Aoyama, A; Noguchi-Yachide, T; Makishima, M; Miyachi, H; Hashimoto, Y Co-existence of alpha-glucosidase-inhibitory and liver X receptor-regulatory activities and their separation by structural development. Bioorg Med Chem 16:4272-85 (2008) [PubMed] Article
Dodo, K; Aoyama, A; Noguchi-Yachide, T; Makishima, M; Miyachi, H; Hashimoto, Y Co-existence of alpha-glucosidase-inhibitory and liver X receptor-regulatory activities and their separation by structural development. Bioorg Med Chem 16:4272-85 (2008) [PubMed] Article More Info.:
Target
Name:
Alpha-glucosidase MAL32
Synonyms:
Alpha-glucosidase MAL32 | MAL32 | MAL32_YEAST | MAL3S | Maltase | alpha-Glucosidase (α-Glucosidase) | alpha-Glucosidase (alpha-Glu) | alpha-Glucosidase (α-Glucosidase)
Type:
Hydrolase
Mol. Mass.:
68132.36
Organism:
Saccharomyces cerevisiae
Description:
Purchased from Wako Fine Chemicals Co. Ltd.
Residue:
584
Sequence:
MTISDHPETEPKWWKEATIYQIYPASFKDSNNDGWGDLKGITSKLQYIKDLGVDAIWVCPFYDSPQQDMGYDISNYEKVWPTYGTNEDCFELIDKTHKLGMKFITDLVINHCSTEHEWFKESRSSKTNPKRDWFFWRPPKGYDAEGKPIPPNNWKSFFGGSAWTFDETTNEFYLRLFASRQVDLNWENEDCRRAIFESAVGFWLDHGVDGFRIDTAGLYSKRPGLPDSPIFDKTSKLQHPNWGSHNGPRIHEYHQELHRFMKNRVKDGREIMTVGEVAHGSDNALYTSAARYEVSEVFSFTHVELGTSPFFRYNIVPFTLKQWKEAIASNFLFINGTDSWATTYIENHDQARSITRFADDSPKYRKISGKLLTLLECSLTGTLYVYQGQEIGQINFKEWPIEKYEDVDVKNNYEIIKKSFGKNSKEMKDFFKGIALLSRDHSRTPMPWTKDKPNAGFTGPDVKPWFFLNESFEQGINVEQESRDDDSVLNFWKRALQARKKYKELMIYGYDFQFIDLDSDQIFSFTKEYEDKTLFAALNFSGEEIEFSLPREGASLSFILGNYDDTDVSSRVLKPWEGRIYLVK
Inhibitor
Name:
BDBM23838
Synonyms:
(2R,5S,14S,16R)-14-[(2S,3R)-3-hydroxy-6-methylheptan-2-yl]-2,16-dimethyltetracyclo[8.7.0.0^{2,7}.0^{11,15}]heptadec-7-en-5-ol | 22-(R)-hydroxycholesterol
Type:
Steroid
Emp. Form.:
C27H46O2
Mol. Mass.:
402.6529
SMILES:
[H][C@@]1(CCC2C1[C@H](C)CC1C2CC=C2C[C@@H](O)CC[C@]12C)[C@H](C)[C@H](O)CCC(C)C |r,t:14|
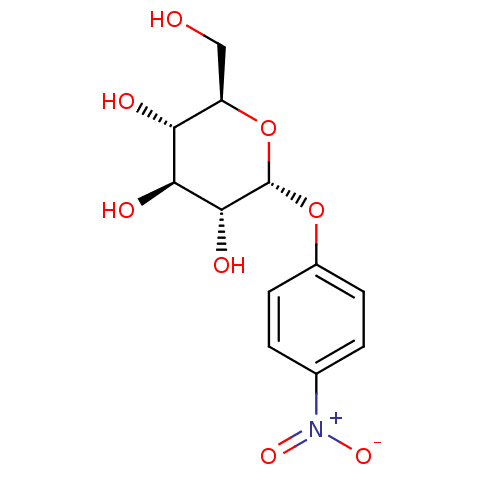
Substrate
Name:
BDBM23837
Synonyms:
(2R,3S,4S,5R,6R)-2-(hydroxymethyl)-6-(4-nitrophenoxy)oxane-3,4,5-triol | 4-nitrophenyl alpha-glucoside | p-NPG | p-Nitrophenyl alpha-D-glucopyranoside | pNPG
Type:
Chromogenic substrate
Emp. Form.:
C12H15NO8
Mol. Mass.:
301.2494
SMILES:
OC[C@H]1O[C@H](Oc2ccc(cc2)[N+]([O-])=O)[C@H](O)[C@@H](O)[C@@H]1O