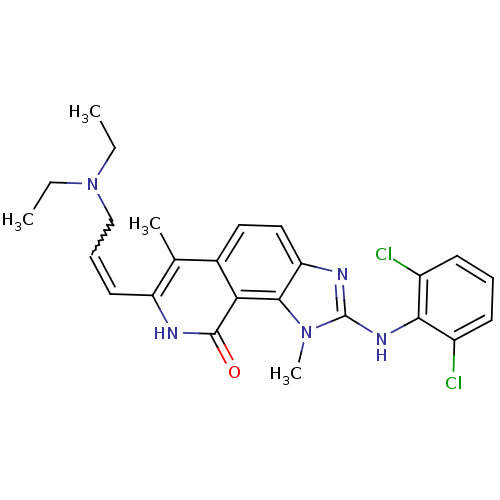
Reaction Details Report a problem with these data
Report a problem with these data
 Report a problem with these data
Report a problem with these dataTarget
Tyrosine-protein kinase ZAP-70
Ligand
BDBM50126732
Substrate
n/a
Meas. Tech.
ChEMBL_217773 (CHEMBL824035)
IC50
400±n/a nM
Citation
 Goldberg, DR; Butz, T; Cardozo, MG; Eckner, RJ; Hammach, A; Huang, J; Jakes, S; Kapadia, S; Kashem, M; Lukas, S; Morwick, TM; Panzenbeck, M; Patel, U; Pav, S; Peet, GW; Peterson, JD; Prokopowicz, AS; Snow, RJ; Sellati, R; Takahashi, H; Tan, J; Tschantz, MA; Wang, XJ; Wang, Y; Wolak, J; Xiong, P; Moss, N Optimization of 2-phenylaminoimidazo[4,5-h]isoquinolin-9-ones: orally active inhibitors of lck kinase. J Med Chem 46:1337-49 (2003) [PubMed] Article
Goldberg, DR; Butz, T; Cardozo, MG; Eckner, RJ; Hammach, A; Huang, J; Jakes, S; Kapadia, S; Kashem, M; Lukas, S; Morwick, TM; Panzenbeck, M; Patel, U; Pav, S; Peet, GW; Peterson, JD; Prokopowicz, AS; Snow, RJ; Sellati, R; Takahashi, H; Tan, J; Tschantz, MA; Wang, XJ; Wang, Y; Wolak, J; Xiong, P; Moss, N Optimization of 2-phenylaminoimidazo[4,5-h]isoquinolin-9-ones: orally active inhibitors of lck kinase. J Med Chem 46:1337-49 (2003) [PubMed] Article More Info.:
Target
Name:
Tyrosine-protein kinase ZAP-70
Synonyms:
70 kDa zeta-associated protein | SRK | Syk-related tyrosine kinase | Tyrosine Kinase ZAP-70 | Tyrosine-protein kinase ZAP-70 | Tyrosine-protein kinase ZAP-70 (Syk) | Tyrosine-protein kinase ZAP-70 (ZAP70) | Tyrosine-protein kinase ZAP70 | ZAP70 | ZAP70_HUMAN | Zeta-chain (TCR) associated protein kinase 70kDa
Type:
Enzyme
Mol. Mass.:
69881.61
Organism:
Homo sapiens (Human)
Description:
ZAP-70 SH2 domain was expressed and purified from E. coli using O-phospho-L-tyrosine-agarose column.
Residue:
619
Sequence:
MPDPAAHLPFFYGSISRAEAEEHLKLAGMADGLFLLRQCLRSLGGYVLSLVHDVRFHHFPIERQLNGTYAIAGGKAHCGPAELCEFYSRDPDGLPCNLRKPCNRPSGLEPQPGVFDCLRDAMVRDYVRQTWKLEGEALEQAIISQAPQVEKLIATTAHERMPWYHSSLTREEAERKLYSGAQTDGKFLLRPRKEQGTYALSLIYGKTVYHYLISQDKAGKYCIPEGTKFDTLWQLVEYLKLKADGLIYCLKEACPNSSASNASGAAAPTLPAHPSTLTHPQRRIDTLNSDGYTPEPARITSPDKPRPMPMDTSVYESPYSDPEELKDKKLFLKRDNLLIADIELGCGNFGSVRQGVYRMRKKQIDVAIKVLKQGTEKADTEEMMREAQIMHQLDNPYIVRLIGVCQAEALMLVMEMAGGGPLHKFLVGKREEIPVSNVAELLHQVSMGMKYLEEKNFVHRDLAARNVLLVNRHYAKISDFGLSKALGADDSYYTARSAGKWPLKWYAPECINFRKFSSRSDVWSYGVTMWEALSYGQKPYKKMKGPEVMAFIEQGKRMECPPECPPELYALMSDCWIYKWEDRPDFLTVEQRMRACYYSLASKVEGPPGSTQKAEAACA
Inhibitor
Name:
BDBM50126732
Synonyms:
2-(2,6-Dichloro-phenylamino)-7-(3-diethylamino-propenyl)-1,6-dimethyl-1,8-dihydro-imidazo[4,5-h]isoquinolin-9-one | CHEMBL281957
Type:
Small organic molecule
Emp. Form.:
C25H27Cl2N5O
Mol. Mass.:
484.421
SMILES:
CCN(CC)CC=Cc1[nH]c(=O)c2c3n(C)c(Nc4c(Cl)cccc4Cl)nc3ccc2c1C |w:6.5|