Reaction Details Report a problem with these data
Report a problem with these data
 Report a problem with these data
Report a problem with these dataTarget
RAF proto-oncogene serine/threonine-protein kinase
Ligand
BDBM50096331
Substrate
n/a
Meas. Tech.
ChEMBL_1495599 (CHEMBL3578337)
IC50
5.2±n/a nM
Citation
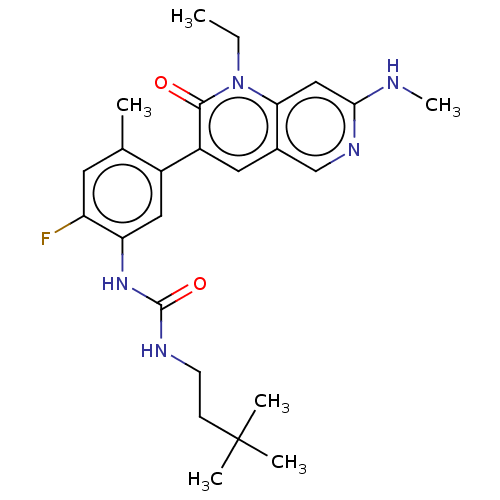
 Henry, JR; Kaufman, MD; Peng, SB; Ahn, YM; Caldwell, TM; Vogeti, L; Telikepalli, H; Lu, WP; Hood, MM; Rutkoski, TJ; Smith, BD; Vogeti, S; Miller, D; Wise, SC; Chun, L; Zhang, X; Zhang, Y; Kays, L; Hipskind, PA; Wrobleski, AD; Lobb, KL; Clay, JM; Cohen, JD; Walgren, JL; McCann, D; Patel, P; Clawson, DK; Guo, S; Manglicmot, D; Groshong, C; Logan, C; Starling, JJ; Flynn, DL Discovery of 1-(3,3-dimethylbutyl)-3-(2-fluoro-4-methyl-5-(7-methyl-2-(methylamino)pyrido[2,3-d]pyrimidin-6-yl)phenyl)urea (LY3009120) as a pan-RAF inhibitor with minimal paradoxical activation and activity against BRAF or RAS mutant tumor cells. J Med Chem 58:4165-79 (2015) [PubMed] Article
Henry, JR; Kaufman, MD; Peng, SB; Ahn, YM; Caldwell, TM; Vogeti, L; Telikepalli, H; Lu, WP; Hood, MM; Rutkoski, TJ; Smith, BD; Vogeti, S; Miller, D; Wise, SC; Chun, L; Zhang, X; Zhang, Y; Kays, L; Hipskind, PA; Wrobleski, AD; Lobb, KL; Clay, JM; Cohen, JD; Walgren, JL; McCann, D; Patel, P; Clawson, DK; Guo, S; Manglicmot, D; Groshong, C; Logan, C; Starling, JJ; Flynn, DL Discovery of 1-(3,3-dimethylbutyl)-3-(2-fluoro-4-methyl-5-(7-methyl-2-(methylamino)pyrido[2,3-d]pyrimidin-6-yl)phenyl)urea (LY3009120) as a pan-RAF inhibitor with minimal paradoxical activation and activity against BRAF or RAS mutant tumor cells. J Med Chem 58:4165-79 (2015) [PubMed] Article More Info.:
Target
Name:
RAF proto-oncogene serine/threonine-protein kinase
Synonyms:
C-Raf Protein Kinase | Proto-oncogene c-RAF (RAF1) | RAF | RAF proto-oncogene
serine/threonine-protein kinase (C-Raf) | RAF1 | RAF1_HUMAN | Raf-1 | Serine/threonine-protein kinase
RAF | Serine/threonine-protein kinase C-Raf | cRaf
Type:
Serine/threonine-protein kinase
Mol. Mass.:
73082.52
Organism:
Homo sapiens (Human)
Description:
P04049
Residue:
648
Sequence:
MEHIQGAWKTISNGFGFKDAVFDGSSCISPTIVQQFGYQRRASDDGKLTDPSKTSNTIRVFLPNKQRTVVNVRNGMSLHDCLMKALKVRGLQPECCAVFRLLHEHKGKKARLDWNTDAASLIGEELQVDFLDHVPLTTHNFARKTFLKLAFCDICQKFLLNGFRCQTCGYKFHEHCSTKVPTMCVDWSNIRQLLLFPNSTIGDSGVPALPSLTMRRMRESVSRMPVSSQHRYSTPHAFTFNTSSPSSEGSLSQRQRSTSTPNVHMVSTTLPVDSRMIEDAIRSHSESASPSALSSSPNNLSPTGWSQPKTPVPAQRERAPVSGTQEKNKIRPRGQRDSSYYWEIEASEVMLSTRIGSGSFGTVYKGKWHGDVAVKILKVVDPTPEQFQAFRNEVAVLRKTRHVNILLFMGYMTKDNLAIVTQWCEGSSLYKHLHVQETKFQMFQLIDIARQTAQGMDYLHAKNIIHRDMKSNNIFLHEGLTVKIGDFGLATVKSRWSGSQQVEQPTGSVLWMAPEVIRMQDNNPFSFQSDVYSYGIVLYELMTGELPYSHINNRDQIIFMVGRGYASPDLSKLYKNCPKAMKRLVADCVKKVKEERPLFPQILSSIELLQHSLPKINRSASEPSLHRAAHTEDINACTLTTSPRLPVF