Reaction Details Report a problem with these data
Report a problem with these data
 Report a problem with these data
Report a problem with these dataTarget
Substance-P receptor
Ligand
BDBM50001450
Substrate
n/a
Ki
1.02±n/a nM
Comments
PDSP_1775
Citation
 Kudlacz, EM; Shatzer, SA; Knippenberg, RW; Logan, DE; Poirot, M; van Giersbergen, PL; Burkholder, TP In vitro and in vivo characterization of MDL 105,212A, a nonpeptide NK-1/NK-2 tachykinin receptor antagonist. J Pharmacol Exp Ther 277:840-51 (1996) [PubMed]
Kudlacz, EM; Shatzer, SA; Knippenberg, RW; Logan, DE; Poirot, M; van Giersbergen, PL; Burkholder, TP In vitro and in vivo characterization of MDL 105,212A, a nonpeptide NK-1/NK-2 tachykinin receptor antagonist. J Pharmacol Exp Ther 277:840-51 (1996) [PubMed] More Info.:
Target
Name:
Substance-P receptor
Synonyms:
NK-1 receptor | NK-1R | NK1 Receptor | NK1R_RAT | Neurokinin 1 receptor | Neurokinin NK1 | SPR | Substance-P receptor | Tac1r | Tachykinin receptor 1 | Tacr1
Type:
G Protein-Coupled Receptor (GPCR)
Mol. Mass.:
46371.54
Organism:
Rattus norvegicus (rat)
Description:
Competition binding assays were carried out using membrane preparations from transfected CHO cells that constitutively expressed the rat NK1 receptor.
Residue:
407
Sequence:
MDNVLPMDSDLFPNISTNTSESNQFVQPTWQIVLWAAAYTVIVVTSVVGNVVVIWIILAHKRMRTVTNYFLVNLAFAEACMAAFNTVVNFTYAVHNVWYYGLFYCKFHNFFPIAALFASIYSMTAVAFDRYMAIIHPLQPRLSATATKVVIFVIWVLALLLAFPQGYYSTTETMPSRVVCMIEWPEHPNRTYEKAYHICVTVLIYFLPLLVIGYAYTVVGITLWASEIPGDSSDRYHEQVSAKRKVVKMMIVVVCTFAICWLPFHVFFLLPYINPDLYLKKFIQQVYLASMWLAMSSTMYNPIIYCCLNDRFRLGFKHAFRCCPFISAGDYEGLEMKSTRYLQTQSSVYKVSRLETTISTVVGAHEEEPEEGPKATPSSLDLTSNGSSRSNSKTMTESSSFYSNMLA
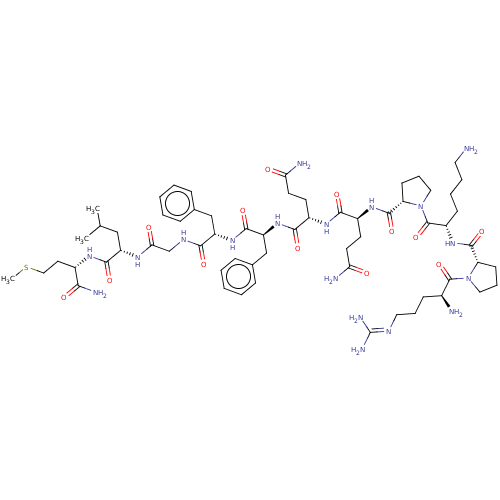
Inhibitor
Name:
BDBM50001450
Synonyms:
(SP)Arg-Pro-Lys-Pro-Gln-Gln-Phe-Phe-Gly-Leu-Met-NH2 | Arg-Pro-Lys-Pro-Gln-Gln-Phe-Phe-Gly-Leu-Met NH2 | Arg-Pro-Lys-Pro-Gln-Gln-Phe-Phe-Gly-Leu-Met-NH2 | Arg-Pro-Lys-Pro-Gln-Gln-Phe-Phe-Gly-Leu-Met-NH2(Substance P) | Arg-Pro-Lys-Pro-Gln-Gln-Phe-Phe-Gly-Leu-Met-NH2.(Substance P) | Arg-Pro-Lys-Pro-Gln-Gln-Phe-Phe-Gly-Leu-Met-amine | ArgProLysProGlnGlnPhePheGlyLeuMet | CHEMBL235363 | H-Arg-Pro-Lys-Pro-Gln-Gln-Phe-D-Phe-Gly-Leu-Met-NH2 | H-Arg-Pro-Lys-Pro-Gln-Gln-Phe-Phe-Gly-Leu-Met-NH2(Substance P) | Substance P | Substance P (Arg-Pro-Lys-Pro-Gln-Gln-Phe-Phe-Gly-Leu-MetNH2) | Substance P analogue | tachykinin substance P (SP)
Type:
Small organic molecule
Emp. Form.:
C63H98N18O13S
Mol. Mass.:
1347.63
SMILES:
[#6]-[#16]-[#6]-[#6]-[#6@H](-[#7]-[#6](=O)-[#6@H](-[#6]-[#6](-[#6])-[#6])-[#7]-[#6](=O)-[#6]-[#7]-[#6](=O)-[#6@H](-[#6]-c1ccccc1)-[#7]-[#6](=O)-[#6@H](-[#6]-c1ccccc1)-[#7]-[#6](=O)-[#6@H](-[#6]-[#6]-[#6](-[#7])=O)-[#7]-[#6](=O)-[#6@H](-[#6]-[#6]-[#6](-[#7])=O)-[#7]-[#6](=O)-[#6@@H]-1-[#6]-[#6]-[#6]-[#7]-1-[#6](=O)-[#6@H](-[#6]-[#6]-[#6]-[#6]-[#7])-[#7]-[#6](=O)-[#6@@H]-1-[#6]-[#6]-[#6]-[#7]-1-[#6](=O)-[#6@@H](-[#7])-[#6]-[#6]-[#6]\[#7]=[#6](\[#7])-[#7])-[#6](-[#7])=O |r|