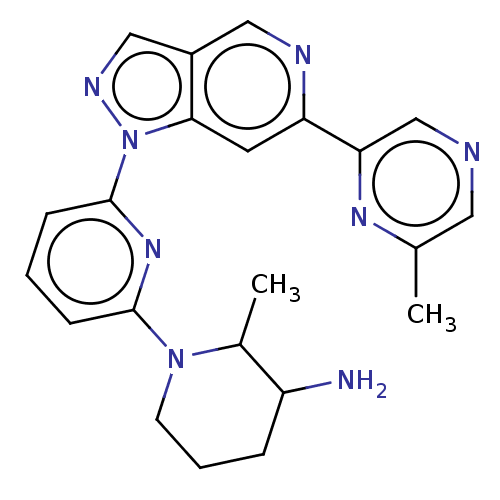
BDBM248964 US9434725, 195::US9434725, 203::US9434725, 204
SMILES CC1C(N)CCCN1c1cccc(n1)-n1ncc2cnc(cc12)-c1cncc(C)n1
InChI Key InChIKey=AVOZYOVWQGVLHZ-UHFFFAOYSA-N
Activity Spreadsheet -- Enzyme Inhibition Constant Data from BindingDB
 Found 12 hits for monomerid = 248964
Found 12 hits for monomerid = 248964
Affinity DataKi: 0.0250nM ΔG°: -14.5kcal/molepH: 7.2 T: 2°CAssay Description:PIM-1, -2, and -3 enzymes were generated as fusion proteins expressed in bacteria and purified by IMAC column chromatography (Sun, X., Chiu, J. F., a...More data for this Ligand-Target Pair
Affinity DataKi: 0.0340nM ΔG°: -14.3kcal/molepH: 7.2 T: 2°CAssay Description:PIM-1, -2, and -3 enzymes were generated as fusion proteins expressed in bacteria and purified by IMAC column chromatography (Sun, X., Chiu, J. F., a...More data for this Ligand-Target Pair
Affinity DataKi: 0.0570nM ΔG°: -14.0kcal/molepH: 7.2 T: 2°CAssay Description:PIM-1, -2, and -3 enzymes were generated as fusion proteins expressed in bacteria and purified by IMAC column chromatography (Sun, X., Chiu, J. F., a...More data for this Ligand-Target Pair
Affinity DataKi: 0.0650nM ΔG°: -13.9kcal/molepH: 7.2 T: 2°CAssay Description:PIM-1, -2, and -3 enzymes were generated as fusion proteins expressed in bacteria and purified by IMAC column chromatography (Sun, X., Chiu, J. F., a...More data for this Ligand-Target Pair
Affinity DataKi: 0.331nM ΔG°: -12.9kcal/molepH: 7.2 T: 2°CAssay Description:PIM-1, -2, and -3 enzymes were generated as fusion proteins expressed in bacteria and purified by IMAC column chromatography (Sun, X., Chiu, J. F., a...More data for this Ligand-Target Pair
Affinity DataKi: 0.375nM ΔG°: -12.9kcal/molepH: 7.2 T: 2°CAssay Description:PIM-1, -2, and -3 enzymes were generated as fusion proteins expressed in bacteria and purified by IMAC column chromatography (Sun, X., Chiu, J. F., a...More data for this Ligand-Target Pair
Affinity DataKi: 0.477nM ΔG°: -12.7kcal/molepH: 7.2 T: 2°CAssay Description:PIM-1, -2, and -3 enzymes were generated as fusion proteins expressed in bacteria and purified by IMAC column chromatography (Sun, X., Chiu, J. F., a...More data for this Ligand-Target Pair
Affinity DataKi: 1.03nM ΔG°: -12.3kcal/molepH: 7.2 T: 2°CAssay Description:PIM-1, -2, and -3 enzymes were generated as fusion proteins expressed in bacteria and purified by IMAC column chromatography (Sun, X., Chiu, J. F., a...More data for this Ligand-Target Pair
Affinity DataKi: 4.83nM ΔG°: -11.3kcal/molepH: 7.2 T: 2°CAssay Description:PIM-1, -2, and -3 enzymes were generated as fusion proteins expressed in bacteria and purified by IMAC column chromatography (Sun, X., Chiu, J. F., a...More data for this Ligand-Target Pair
Affinity DataIC50: 693nMAssay Description:BaF3, a murine interleukin-3 dependent pro-B cell line, parental cells, BaF3 PIM1 cells, BaF3 PIM2 cells, and MM1.S (multiple myeloma) cells were see...More data for this Ligand-Target Pair
Affinity DataIC50: 1.00E+3nMAssay Description:BaF3, a murine interleukin-3 dependent pro-B cell line, parental cells, BaF3 PIM1 cells, BaF3 PIM2 cells, and MM1.S (multiple myeloma) cells were see...More data for this Ligand-Target Pair
Affinity DataIC50: 2.00E+3nMAssay Description:BaF3, a murine interleukin-3 dependent pro-B cell line, parental cells, BaF3 PIM1 cells, BaF3 PIM2 cells, and MM1.S (multiple myeloma) cells were see...More data for this Ligand-Target Pair