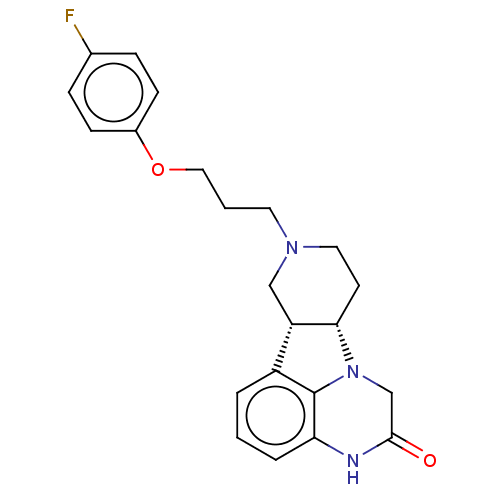
BDBM683053 US12023331, Example 1::US20240307386, Compound Example 1
SMILES Fc1ccc(OCCCN2CC[C@H]3[C@@H](C2)c2cccc4NC(=O)CN3c24)cc1
InChI Key InChIKey=MXIJXDUPULMKIE-UHFFFAOYSA-N
Data 10 KI
Activity Spreadsheet -- Enzyme Inhibition Constant Data from BindingDB
 Found 25 hits for monomerid = 683053
Found 25 hits for monomerid = 683053
Affinity DataKi: 8.30nMAssay Description:Receptor binding is determined for the Compounds of Examples 1, 2 and 3 (corresponding to Formula 1, Formula B and Formula A, respectively). The foll...More data for this Ligand-Target Pair
Affinity DataKi: 8.30nMAssay Description:Receptor binding is determined for the Compound of Example 1, using the compound of Formula A as a control. The following literature procedures are u...More data for this Ligand-Target Pair
Affinity DataKi: 8.30nMAssay Description:In general, the results are expressed as a percent of control specific binding:measuredspecificbindingcontrolspecificbinding×100and as a percent ...More data for this Ligand-Target Pair
Affinity DataKi: 8.30nMAssay Description:Displacement of [125I]DOI from human 5-HT2A receptor expressed in HEK293 cell membrane assessed as inhibition constant incubated for 60 mins by compe...More data for this Ligand-Target Pair
Affinity DataKi: 8.30nMAssay Description:In general, the results are expressed as a percent of control specific binding: measuredspecificbindingcontrolspecificbin...More data for this Ligand-Target Pair
Affinity DataKi: 11nMAssay Description:Receptor binding is determined for the Compound of Example 1, using the compound of Formula A as a control. The following literature procedures are u...More data for this Ligand-Target Pair
Affinity DataKi: 11nMAssay Description:In general, the results are expressed as a percent of control specific binding: measuredspecificbindingcontrolspecificbin...More data for this Ligand-Target Pair
Affinity DataKi: 11nMAssay Description:In general, the results are expressed as a percent of control specific binding:measuredspecificbindingcontrolspecificbinding×100and as a percent ...More data for this Ligand-Target Pair
Affinity DataKi: 11nMAssay Description:Displacement of [3H]DAMGO from human mu opioid receptor expressed in HEK293 cell membrane assessed as inhibition constant incubated for 120 mins by c...More data for this Ligand-Target Pair
Affinity DataKi: 11nMAssay Description:Receptor binding is determined for the Compounds of Examples 1, 2 and 3 (corresponding to Formula 1, Formula B and Formula A, respectively). The foll...More data for this Ligand-Target Pair
Affinity DataKi: 50nMAssay Description:Receptor binding is determined for the Compounds of Examples 1, 2 and 3 (corresponding to Formula 1, Formula B and Formula A, respectively). The foll...More data for this Ligand-Target Pair
Affinity DataKi: 50nMAssay Description:Receptor binding is determined for the Compound of Example 1, using the compound of Formula A as a control. The following literature procedures are u...More data for this Ligand-Target Pair
Affinity DataKi: 50nMAssay Description:In general, the results are expressed as a percent of control specific binding:measuredspecificbindingcontrolspecificbinding×100and as a percent ...More data for this Ligand-Target Pair
Affinity DataKi: 50nMAssay Description:In general, the results are expressed as a percent of control specific binding: measuredspecificbindingcontrolspecificbin...More data for this Ligand-Target Pair
Affinity DataKi: 50nMAssay Description:Displacement of [3H]SCH-23390 from human dopamine D1 receptor expressed in CHO cell membrane assessed as inhibition constant incubated for 60 mins by...More data for this Ligand-Target Pair
Affinity DataKi: 160nMAssay Description:Receptor binding is determined for the Compounds of Examples 1, 2 and 3 (corresponding to Formula 1, Formula B and Formula A, respectively). The foll...More data for this Ligand-Target Pair
Affinity DataKi: 160nMAssay Description:In general, the results are expressed as a percent of control specific binding:measuredspecificbindingcontrolspecificbinding×100and as a percent ...More data for this Ligand-Target Pair
Affinity DataKi: 160nMAssay Description:Displacement of [3H]7-OH-DPAT from human dopamine D2 receptor expressed in HEK293 cell membrane assessed as inhibition constant incubated for 60 mins...More data for this Ligand-Target Pair
Affinity DataKi: 160nMAssay Description:Receptor binding is determined for the Compound of Example 1, using the compound of Formula A as a control. The following literature procedures are u...More data for this Ligand-Target Pair
Affinity DataKi: 160nMAssay Description:In general, the results are expressed as a percent of control specific binding: measuredspecificbindingcontrolspecificbin...More data for this Ligand-Target Pair
Affinity DataKi: 590nMAssay Description:Receptor binding is determined for the Compound of Example 1, using the compound of Formula A as a control. The following literature procedures are u...More data for this Ligand-Target Pair
Affinity DataKi: 590nMAssay Description:In general, the results are expressed as a percent of control specific binding:measuredspecificbindingcontrolspecificbinding×100and as a percent ...More data for this Ligand-Target Pair
Affinity DataKi: 590nMAssay Description:Displacement of [3H]-imipramine from human 5-HT transporter expressed in CHO cell membrane assessed as inhibition constant incubated for 60 mins by c...More data for this Ligand-Target Pair
Affinity DataKi: 590nMAssay Description:In general, the results are expressed as a percent of control specific binding: measuredspecificbindingcontrolspecificbin...More data for this Ligand-Target Pair
Affinity DataKi: 590nMAssay Description:Receptor binding is determined for the Compounds of Examples 1, 2 and 3 (corresponding to Formula 1, Formula B and Formula A, respectively). The foll...More data for this Ligand-Target Pair