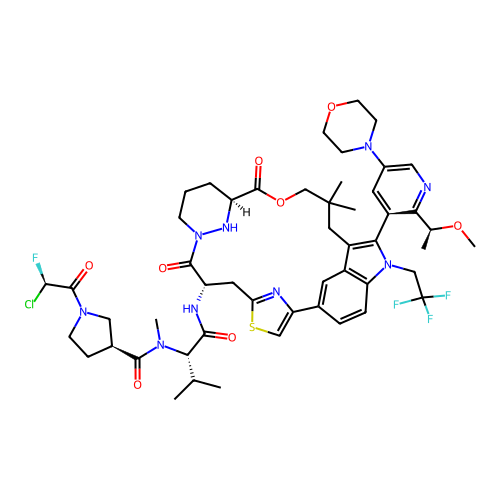
BDBM743469 (3S)-1-[(2R)-2-chloro-2-fluoro-acetyl]-N-[(1S)-1-[[(7S,13S)-(20M)-20-[2-[(1S)-1-methoxyethyl]-5-morpholino-3-pyridyl]-17,17-dimethyl-8,14-dioxo-21-(2,2,2-trifluoroethyl)-15-oxa-4-thia-9,21,27,28-tetrazapentacyclo[17.5.2.12,5.19,13.022,26]octacosa-1(25),2,5(28),19,22::US20250163072, Example 25
SMILES CO[C@@H](C)c1ncc(N2CCOCC2)cc1-c1c2c3cc(ccc3n1CC(F)(F)F)-c1csc(n1)C[C@H](NC(=O)[C@H](C(C)C)N(C)C(=O)[C@H]1CCN(C(=O)[C@H](F)Cl)C1)C(=O)N1CCC[C@H](N1)C(=O)OCC(C)(C)C2
InChI Key
Activity Spreadsheet -- Enzyme Inhibition Constant Data from BindingDB
 Found 4 hits for monomerid = 743469
Found 4 hits for monomerid = 743469
Affinity DataIC50: 20nMAssay Description:HEK293 cells were grown and maintained using DMEM medium (Thermo Fisher Scientific) with 10% fetal bovine serum and 1% penicillin/streptomycin. Both ...More data for this Ligand-Target Pair
Affinity DataIC50: 32nMAssay Description:In this example, TR-FRET was also used to measure the compound or compound-CYPA dependent disruption of the KRAS G12C-BRAF complex. This protocol was...More data for this Ligand-Target Pair
Affinity DataIC50: 1.00E+4nMAssay Description:In this example, TR-FRET was also used to measure the compound or compound-CYPA dependent disruption of the KRAS G12C-BRAF complex. This protocol was...More data for this Ligand-Target Pair
Affinity DataIC50: 1.00E+4nMAssay Description:In this example, TR-FRET was also used to measure the compound or compound-CYPA dependent disruption of the KRAS G12C-BRAF complex. This protocol was...More data for this Ligand-Target Pair