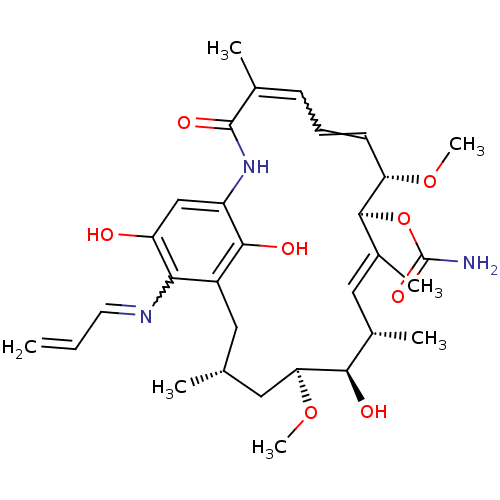
BDBM15359 (4E,6Z,8S,9S,10E,12S,13R,14S,16R)-13-hydroxy-8,14-dimethoxy-4,10,12,16-tetramethyl-3,20,22-trioxo-19-(prop-2-en-1-ylamino)-2-azabicyclo[16.3.1]docosa-1(21),4,6,10,18-pentaen-9-yl carbamate::17-(Allylamino)geldanamycin::17-AAG::17AAG::CHEMBL109480::GLD-36::Tanespimycin
SMILES CO[C@H]1C[C@H](C)Cc2c(O)c(NC(=O)C(C)=CC=C[C@H](OC)[C@@H](OC(N)=O)\C(C)=C\[C@H](C)[C@H]1O)cc(O)c2N=CC=C
InChI Key InChIKey=VLKLQQLCVPDQFU-UHFFFAOYSA-N
Activity Spreadsheet -- Enzyme Inhibition Constant Data from BindingDB
 Found 41 hits for monomerid = 15359
Found 41 hits for monomerid = 15359
Affinity DataKi: 4.60nMAssay Description:Inhibition of HSP90alphaMore data for this Ligand-Target Pair
Affinity DataEC50: 7nMpH: 7.2 T: 2°CAssay Description:EC50 was defined as the concentration of the compound at which there was 50% degradation of the Her-2/neu protein in MCF7 breast carcinoma cells. Sam...More data for this Ligand-Target Pair
Affinity DataIC50: 7nMAssay Description:Inhibition of HSP90-mediated client protein HER2 degradationMore data for this Ligand-Target Pair
Affinity DataKd: 7.30nMAssay Description:Binding affinity to human Hsp90beta after overnight incubation by fluorescence polarization assayMore data for this Ligand-Target Pair
Affinity DataKi: 9nMAssay Description:Binding affinity to HSP90 under reducing conditions in presence of TECPMore data for this Ligand-Target Pair
Affinity DataIC50: 12nMAssay Description:Inhibition of HSP90-mediated client protein HER2 degradation in human MCF7 cellsMore data for this Ligand-Target Pair
Affinity DataIC50: 15nMAssay Description:Inhibition of Hsp90 in human MCF7 cells assessed as Her2 degradationMore data for this Ligand-Target Pair
Affinity DataEC50: 19nMAssay Description:Degradation of Her2 in SKOV3 cellsMore data for this Ligand-Target Pair
Affinity DataIC50: 20nMAssay Description:Binding affinity to Hsp90alpha N-terminal ATPase domain by TR-FRET assay based competitive binding assayMore data for this Ligand-Target Pair
Affinity DataEC50: 22nMAssay Description:Upregulation of Hsp70 in SKBR3 cellsMore data for this Ligand-Target Pair
Affinity DataIC50: 31nMAssay Description:Displacement of 5-(3-(3-(6-amino-8-(6-iodobenzo[d][1,3]dioxol-5-ylthio)-9H-purin-9-yl)propyl)thioureido)-2-(6-hydroxy-3-oxo-3H-xanthen-9-yl)benzoic a...More data for this Ligand-Target Pair
TargetReceptor tyrosine-protein kinase erbB-2(Human)
Memorial Sloan-Kettering Cancer Center
Curated by ChEMBL
Memorial Sloan-Kettering Cancer Center
Curated by ChEMBL
Affinity DataIC50: 31nMAssay Description:Inhibition of Her2 in human SKBR3 cellsMore data for this Ligand-Target Pair
Affinity DataIC50: 33nMAssay Description:Binding affinity to Hsp90 in human SKBR3 cellsMore data for this Ligand-Target Pair
Affinity DataEC50: 35nMAssay Description:Degradation of Her2 in SKBR3 cellsMore data for this Ligand-Target Pair
Affinity DataEC50: 45nMAssay Description:Displacement of GM-BODIPY from Hsp90More data for this Ligand-Target Pair
Affinity DataEC50: 60nMAssay Description:Inhibition of Hsp90alpha in human SKBR3 cells assessed as up-regulation of HSP70 protein by Western blot analysisMore data for this Ligand-Target Pair
Affinity DataEC50: 64.9nMAssay Description:Displacement of cy3B-GM from Hsp90alphaMore data for this Ligand-Target Pair
Affinity DataEC50: 110nMAssay Description:Binding affinity towards Heat shock protein HSP 90-alpha inhibitorMore data for this Ligand-Target Pair
Affinity DataEC50: 120nMAssay Description:Inhibition of Hsp90alpha in human H1299 cells assessed as HIF-1alpha degradation by luciferase reporter assayMore data for this Ligand-Target Pair
Affinity DataEC50: 124nMAssay Description:Inhibition of BODIPY-AG binding to dog Grp94More data for this Ligand-Target Pair
Affinity DataKd: 160nMAssay Description:Binding affinity to human recombinant Hsp90alpha N-terminal domain by isothermal titration calorimetryMore data for this Ligand-Target Pair
Affinity DataIC50: 1.27E+3nM EC50: 160nMpH: 7.4 T: 2°CAssay Description:The assay is based upon displacement of a fluorescently labeled molecule, which binds specifically to the ATP-binding site of full-length human Hsp90...More data for this Ligand-Target Pair
Affinity DataKd: 220nMAssay Description:Binding affinity to biotinylated N-terminal human HSP90alpha using of streptavidin coated surface by SPR binding assayMore data for this Ligand-Target Pair
Affinity DataKd: 220nMAssay Description:Binding affinity to human HSP90alpha assessed as 2D1H-15N chemical shift perturbation by NMR spectroscopyMore data for this Ligand-Target Pair
Affinity DataKd: 326nMAssay Description:Binding affinity to recombinant Hsp90 alpha (unknown origin) by surface plasmon resonanceMore data for this Ligand-Target Pair
Affinity DataKi: 530nMAssay Description:Binding affinity to HSP90 under non-reducing conditions in absence of TECPMore data for this Ligand-Target Pair
Affinity DataKd: 760nMAssay Description:Binding affinity to human recombinant Hsp90alpha N-terminal domain by scintillation proximity assayMore data for this Ligand-Target Pair
Affinity DataEC50: 1.00E+3nMAssay Description:Competitive binding towards Heat shock protein HSP 90-alphaMore data for this Ligand-Target Pair
Affinity DataEC50: 1.00E+3nMAssay Description:Inhibition of Hsp90More data for this Ligand-Target Pair
Affinity DataIC50: 1.19E+3nMAssay Description:Displacement of FITC-labeled geldanamycin from human recombinant HSP90alpha after 24 hrs by fluorescence displacement assayMore data for this Ligand-Target Pair
Affinity DataIC50: 1.27E+3nMpH: 7.4 T: 2°CAssay Description:The assay is based upon displacement of a fluorescently labeled molecule, which binds specifically to the ATP-binding site of full-length human Hsp90...More data for this Ligand-Target Pair
Affinity DataIC50: 1.27E+3nMpH: 7.4 T: 2°CAssay Description:The assay is based upon displacement of a fluorescently labeled molecule, which binds specifically to the ATP-binding site of full-length human Hsp90...More data for this Ligand-Target Pair
Affinity DataIC50: 1.27E+3nMAssay Description:Binding affinity to HSP90More data for this Ligand-Target Pair
Affinity DataKd: 1.30E+3nMAssay Description:Binding affinity to human recombinant HSP90More data for this Ligand-Target Pair
TargetHeat shock protein 75 kDa, mitochondrial(Human)
Memorial Sloan-Kettering Cancer Center
Curated by ChEMBL
Memorial Sloan-Kettering Cancer Center
Curated by ChEMBL
Affinity DataIC50: 1.50E+3nMAssay Description:Displacement of 5-(3-(3-(6-amino-8-(6-iodobenzo[d][1,3]dioxol-5-ylthio)-9H-purin-9-yl)propyl)thioureido)-2-(6-hydroxy-3-oxo-3H-xanthen-9-yl)benzoic a...More data for this Ligand-Target Pair
TargetATP-dependent molecular chaperone HSP82(Baker's yeast)
The Institute of Cancer Research
Curated by ChEMBL
The Institute of Cancer Research
Curated by ChEMBL
Affinity DataIC50: 8.70E+3nMAssay Description:Inhibition of yeast Hsp90 ATPase activityMore data for this Ligand-Target Pair
TargetATP-dependent molecular chaperone HSP82(Baker's yeast)
The Institute of Cancer Research
Curated by ChEMBL
The Institute of Cancer Research
Curated by ChEMBL
Affinity DataIC50: 8.70E+3nMAssay Description:Inhibition of yeast Hsp90 ATPase activityMore data for this Ligand-Target Pair
Affinity DataIC50: 9.80E+3nMAssay Description:Inhibition of Hsp90alpha ATPase activity by malachite green ATP-ase assayMore data for this Ligand-Target Pair
Affinity DataIC50: 1.34E+4nMAssay Description:Inhibition of ATP-ase activity in human colon tumor cell line (HCT116)More data for this Ligand-Target Pair
TargetATP-dependent molecular chaperone HSP82(Baker's yeast)
The Institute of Cancer Research
Curated by ChEMBL
The Institute of Cancer Research
Curated by ChEMBL
Affinity DataIC50: 1.34E+4nMpH: 7.4 T: 2°CAssay Description:The HSP90 ATPase activity was determined by following the procedure of malachite green assay. The assay is based on quantitation of the green complex...More data for this Ligand-Target Pair
TargetDNA-dependent protein kinase catalytic subunit(Human)
Department of Molecular Oncogenesis
Curated by ChEMBL
Department of Molecular Oncogenesis
Curated by ChEMBL
Affinity DataIC50: 4.00E+4nMAssay Description:Inhibition of DNA dependent protein kinase activity without hsp90 alpha proteinMore data for this Ligand-Target Pair
Activity Spreadsheet -- ITC Data from BindingDB
 Found 4 hits for monomerid = 15359
Found 4 hits for monomerid = 15359
ITC DataΔG°: -7.74kcal/mole −TΔS°: -3.25kcal/mole ΔH°: -4.61kcal/mole logk: 1.07E+6
pH: 8.0 T: 10.00°C
pH: 8.0 T: 10.00°C
ITC DataΔG°: -7.69kcal/mole −TΔS°: -1.48kcal/mole ΔH°: -6.21kcal/mole logk: 7.50E+5
pH: 8.0 T: 15.00°C
pH: 8.0 T: 15.00°C
ITC DataΔG°: -7.67kcal/mole −TΔS°: -1.17kcal/mole ΔH°: -6.50kcal/mole logk: 5.40E+5
pH: 8.0 T: 20.00°C
pH: 8.0 T: 20.00°C
ITC DataΔG°: -7.62kcal/mole −TΔS°: -0.768kcal/mole ΔH°: -6.85kcal/mole logk: 3.20E+5
pH: 8.0 T: 30.00°C
pH: 8.0 T: 30.00°C