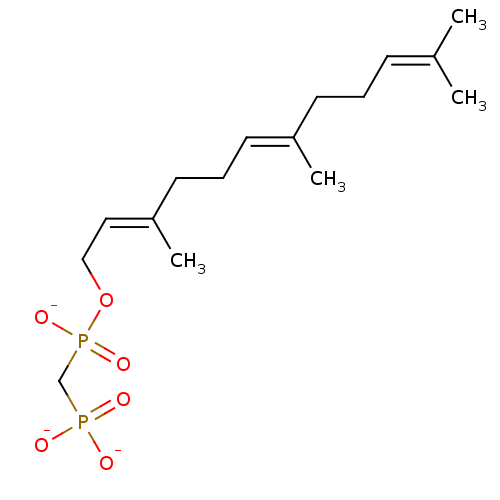
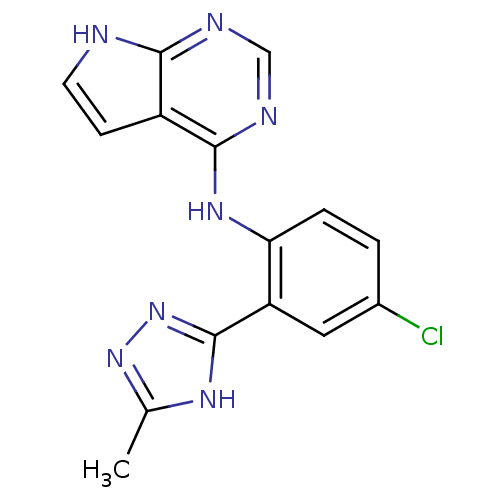
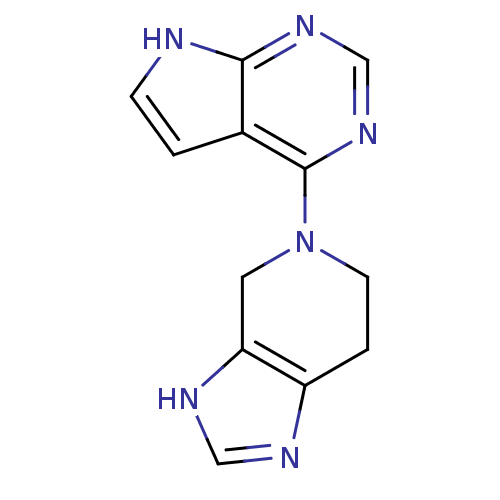
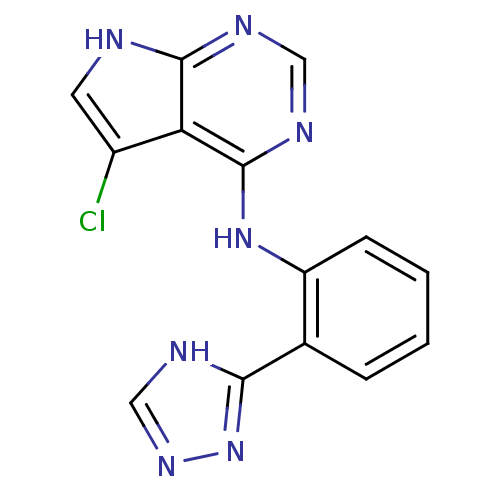
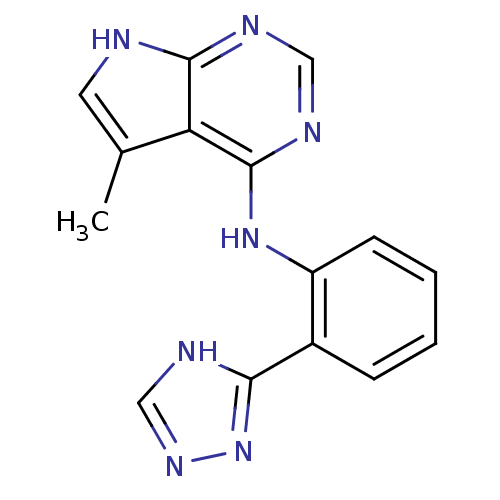
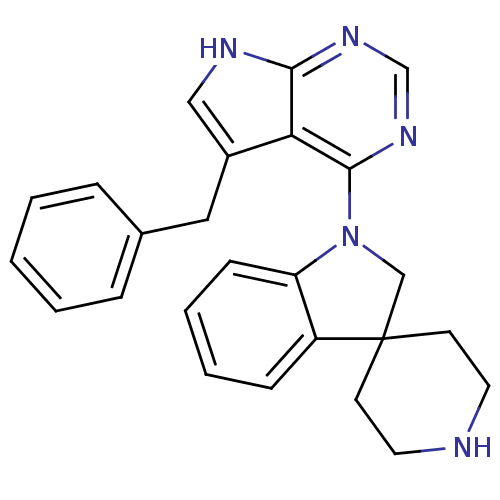
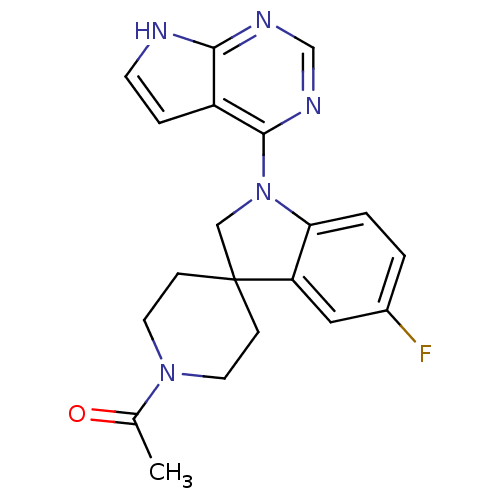
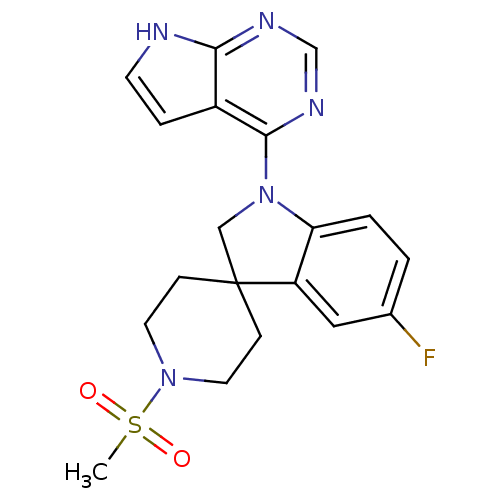
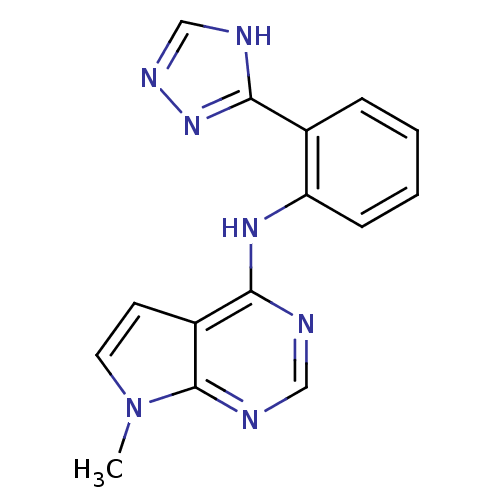
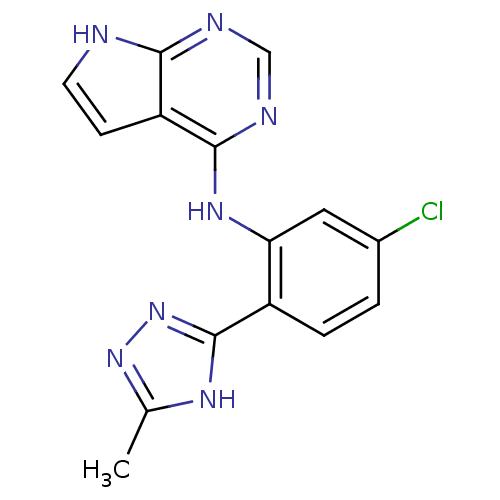
Report error Found 49 with Last Name = 'troutman' and Initial = 'm'
Affinity DataKi: 0.390nMAssay Description:Displacement of [3H]dofetilide from hERG expressed in HEK293 cells by SPAMore data for this Ligand-Target Pair
Affinity DataKi: 2.90nMAssay Description:Binding affinity at hERG expressed in HEK293 cells by fluorescence polarization assayMore data for this Ligand-Target Pair
Affinity DataKi: 3.60nMAssay Description:Displacement of [3H]dofetilide from hERG expressed in HEK293 cells by SPAMore data for this Ligand-Target Pair
Affinity DataKi: 6.40nMAssay Description:Displacement of [3H]dofetilide from hERG expressed in HEK293 cells by SPAMore data for this Ligand-Target Pair
Affinity DataKi: 7nMAssay Description:Displacement of [3H]dofetilide from hERG expressed in HEK293 cells by SPAMore data for this Ligand-Target Pair
Affinity DataKi: 7.70nMAssay Description:Binding affinity at hERG expressed in HEK293 cells by fluorescence polarization assayMore data for this Ligand-Target Pair
Affinity DataKi: 10nMAssay Description:Displacement of [3H]dofetilide from hERG expressed in HEK293 cells by SPAMore data for this Ligand-Target Pair
Affinity DataKi: 13nMAssay Description:Displacement of [3H]dofetilide from hERG expressed in HEK293 cells by SPAMore data for this Ligand-Target Pair
Affinity DataKi: 13nMAssay Description:Displacement of [3H]dofetilide from hERG expressed in HEK293 cells by SPAMore data for this Ligand-Target Pair
Affinity DataKi: 15nMAssay Description:Displacement of [3H]dofetilide from hERG expressed in HEK293 cells by SPAMore data for this Ligand-Target Pair
Affinity DataKi: 18nMAssay Description:Displacement of [3H]dofetilide from hERG expressed in HEK293 cells by SPAMore data for this Ligand-Target Pair
Affinity DataKi: 19nMAssay Description:Displacement of [3H]dofetilide from hERG expressed in HEK293 cells by SPAMore data for this Ligand-Target Pair
Affinity DataKi: 19nMAssay Description:Displacement of [3H]dofetilide from hERG expressed in HEK293 cells by SPAMore data for this Ligand-Target Pair
Affinity DataKi: 19nMAssay Description:Binding affinity at hERG expressed in HEK293 cells by fluorescence polarization assayMore data for this Ligand-Target Pair
Affinity DataKi: 89nMAssay Description:Displacement of [3H]dofetilide from hERG expressed in HEK293 cells by SPAMore data for this Ligand-Target Pair
Affinity DataKi: 160nMAssay Description:Displacement of [3H]dofetilide from hERG expressed in HEK293 cells by SPAMore data for this Ligand-Target Pair
Affinity DataKi: 184nMAssay Description:Binding affinity at hERG expressed in HEK293 cells by fluorescence polarization assayMore data for this Ligand-Target Pair
Affinity DataKi: 190nMAssay Description:Binding affinity at hERG expressed in HEK293 cells by fluorescence polarization assayMore data for this Ligand-Target Pair
Affinity DataKi: 291nMAssay Description:Displacement of [3H]dofetilide from hERG expressed in HEK293 cells by SPAMore data for this Ligand-Target Pair
Affinity DataKi: 353nMAssay Description:Displacement of [3H]dofetilide from hERG expressed in HEK293 cells by SPAMore data for this Ligand-Target Pair
Affinity DataKi: 980nMAssay Description:Displacement of [3H]dofetilide from hERG expressed in HEK293 cells by SPAMore data for this Ligand-Target Pair
Affinity DataKi: >1.00E+3nMAssay Description:Displacement of [3H]dofetilide from hERG expressed in HEK293 cells by SPAMore data for this Ligand-Target Pair
Affinity DataKi: >1.00E+3nMAssay Description:Displacement of [3H]dofetilide from hERG expressed in HEK293 cells by SPAMore data for this Ligand-Target Pair
Affinity DataKi: 3.08E+3nMAssay Description:Displacement of [3H]dofetilide from hERG expressed in HEK293 cells by SPAMore data for this Ligand-Target Pair
Affinity DataKi: 4.06E+3nMAssay Description:Binding affinity at hERG expressed in HEK293 cells by fluorescence polarization assayMore data for this Ligand-Target Pair
Affinity DataKi: 7.30E+3nMAssay Description:Displacement of [3H]dofetilide from hERG expressed in HEK293 cells by SPAMore data for this Ligand-Target Pair
Affinity DataIC50: 2.40nMpH: 7.5 T: 22°CAssay Description:A fluorescence polarization IMAP type assay is used. After a 90 min incubation of reaction mixtures containing enzyme, substrate, and inhibitor, IMAP...More data for this Ligand-Target Pair
Affinity DataIC50: <3nMAssay Description:Blockade of hERG expressed in HEK293 cells by whole-cell patch clamp methodMore data for this Ligand-Target Pair
Affinity DataIC50: 3.10nMpH: 7.5 T: 22°CAssay Description:A fluorescence polarization IMAP type assay is used. After a 90 min incubation of reaction mixtures containing enzyme, substrate, and inhibitor, IMAP...More data for this Ligand-Target Pair
Affinity DataIC50: 4.80nMpH: 7.5 T: 22°CAssay Description:A fluorescence polarization IMAP type assay is used. After a 90 min incubation of reaction mixtures containing enzyme, substrate, and inhibitor, IMAP...More data for this Ligand-Target Pair
Affinity DataIC50: 4.80nMpH: 7.5 T: 22°CAssay Description:A fluorescence polarization IMAP type assay is used. After a 90 min incubation of reaction mixtures containing enzyme, substrate, and inhibitor, IMAP...More data for this Ligand-Target Pair
Affinity DataIC50: 5.30nMpH: 7.5 T: 22°CAssay Description:A fluorescence polarization IMAP type assay is used. After a 90 min incubation of reaction mixtures containing enzyme, substrate, and inhibitor, IMAP...More data for this Ligand-Target Pair
Affinity DataIC50: 5.80nMpH: 7.5 T: 22°CAssay Description:A fluorescence polarization IMAP type assay is used. After a 90 min incubation of reaction mixtures containing enzyme, substrate, and inhibitor, IMAP...More data for this Ligand-Target Pair
Affinity DataIC50: 9.20nMAssay Description:Blockade of hERG expressed in HEK293 cells by whole-cell patch clamp methodMore data for this Ligand-Target Pair
Affinity DataIC50: 12.7nMpH: 7.5 T: 22°CAssay Description:A fluorescence polarization IMAP type assay is used. After a 90 min incubation of reaction mixtures containing enzyme, substrate, and inhibitor, IMAP...More data for this Ligand-Target Pair
Affinity DataIC50: 42nMpH: 7.5 T: 22°CAssay Description:A fluorescence polarization IMAP type assay is used. After a 90 min incubation of reaction mixtures containing enzyme, substrate, and inhibitor, IMAP...More data for this Ligand-Target Pair
Affinity DataIC50: 151nMpH: 7.5 T: 22°CAssay Description:A fluorescence polarization IMAP type assay is used. After a 90 min incubation of reaction mixtures containing enzyme, substrate, and inhibitor, IMAP...More data for this Ligand-Target Pair
TargetProtein farnesyltransferase subunit beta/geranylgeranyltransferase type-1 subunit alpha(Human)
University of Kentucky
Curated by ChEMBL
University of Kentucky
Curated by ChEMBL
Affinity DataIC50: 160nMAssay Description:Inhibition of FarnesyltransferaseMore data for this Ligand-Target Pair
Affinity DataIC50: 171nMpH: 7.5 T: 22°CAssay Description:A fluorescence polarization IMAP type assay is used. After a 90 min incubation of reaction mixtures containing enzyme, substrate, and inhibitor, IMAP...More data for this Ligand-Target Pair
Affinity DataIC50: 210nMpH: 7.5 T: 22°CAssay Description:A fluorescence polarization IMAP type assay is used. After a 90 min incubation of reaction mixtures containing enzyme, substrate, and inhibitor, IMAP...More data for this Ligand-Target Pair
Affinity DataIC50: 294nMpH: 7.5 T: 22°CAssay Description:A fluorescence polarization IMAP type assay is used. After a 90 min incubation of reaction mixtures containing enzyme, substrate, and inhibitor, IMAP...More data for this Ligand-Target Pair
Affinity DataIC50: 310nMAssay Description:Blockade of hERG expressed in HEK293 cells by whole-cell patch clamp methodMore data for this Ligand-Target Pair
Affinity DataIC50: 318nMpH: 7.5 T: 22°CAssay Description:A fluorescence polarization IMAP type assay is used. After a 90 min incubation of reaction mixtures containing enzyme, substrate, and inhibitor, IMAP...More data for this Ligand-Target Pair
Affinity DataIC50: 425nMpH: 7.5 T: 22°CAssay Description:A fluorescence polarization IMAP type assay is used. After a 90 min incubation of reaction mixtures containing enzyme, substrate, and inhibitor, IMAP...More data for this Ligand-Target Pair
Affinity DataIC50: 1.33E+3nMpH: 7.5 T: 22°CAssay Description:A fluorescence polarization IMAP type assay is used. After a 90 min incubation of reaction mixtures containing enzyme, substrate, and inhibitor, IMAP...More data for this Ligand-Target Pair
Affinity DataIC50: 2.79E+3nMpH: 7.5 T: 22°CAssay Description:A fluorescence polarization IMAP type assay is used. After a 90 min incubation of reaction mixtures containing enzyme, substrate, and inhibitor, IMAP...More data for this Ligand-Target Pair
Affinity DataIC50: 3.19E+3nMpH: 7.5 T: 22°CAssay Description:A fluorescence polarization IMAP type assay is used. After a 90 min incubation of reaction mixtures containing enzyme, substrate, and inhibitor, IMAP...More data for this Ligand-Target Pair
Affinity DataIC50: >1.00E+4nMpH: 7.5 T: 22°CAssay Description:A fluorescence polarization IMAP type assay is used. After a 90 min incubation of reaction mixtures containing enzyme, substrate, and inhibitor, IMAP...More data for this Ligand-Target Pair
Affinity DataIC50: >1.00E+4nMpH: 7.5 T: 22°CAssay Description:A fluorescence polarization IMAP type assay is used. After a 90 min incubation of reaction mixtures containing enzyme, substrate, and inhibitor, IMAP...More data for this Ligand-Target Pair
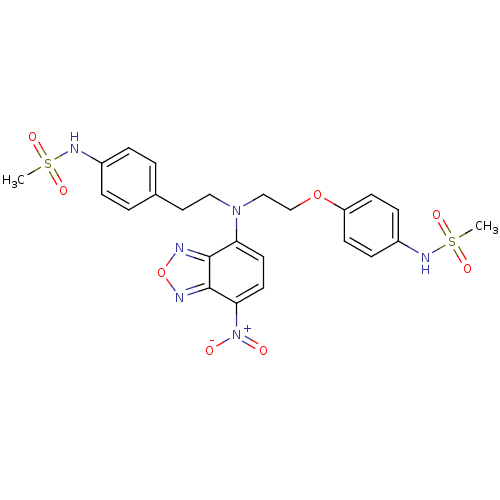
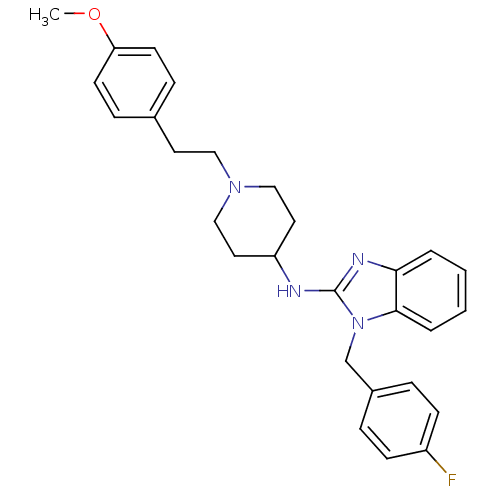
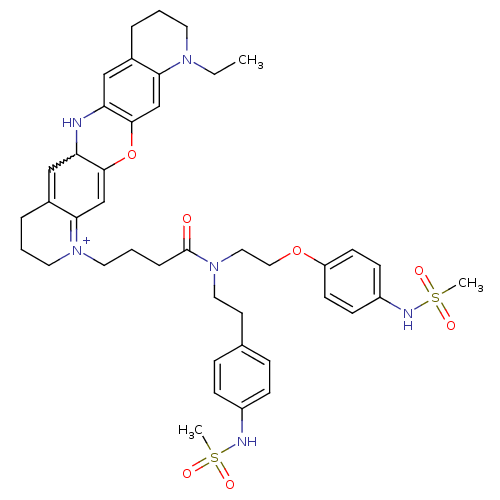
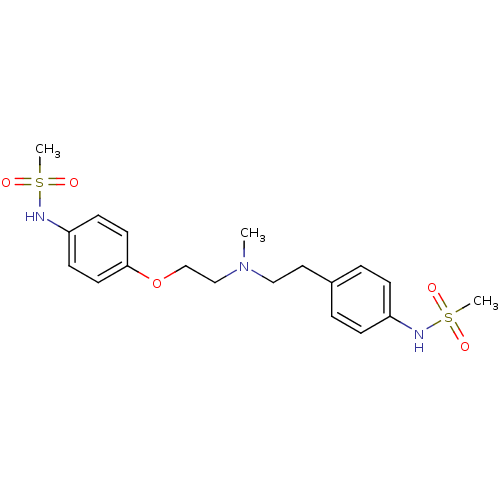
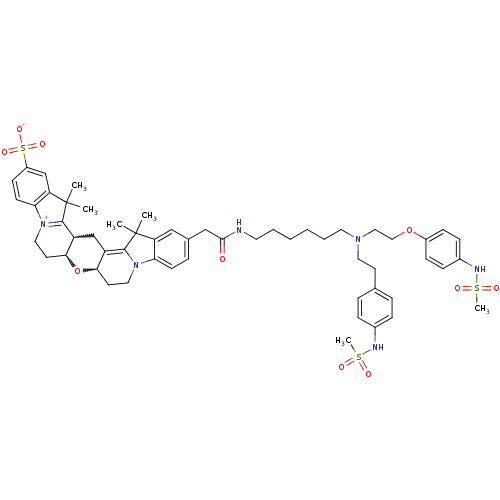
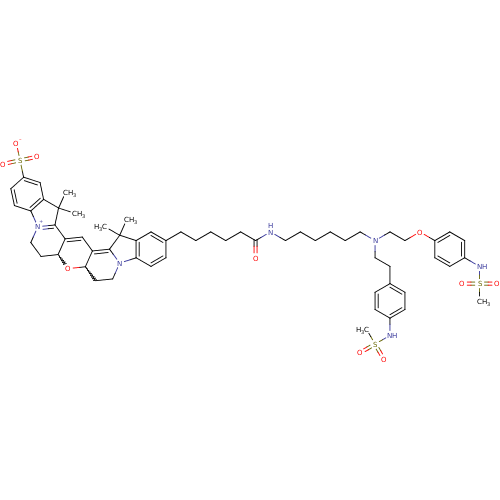
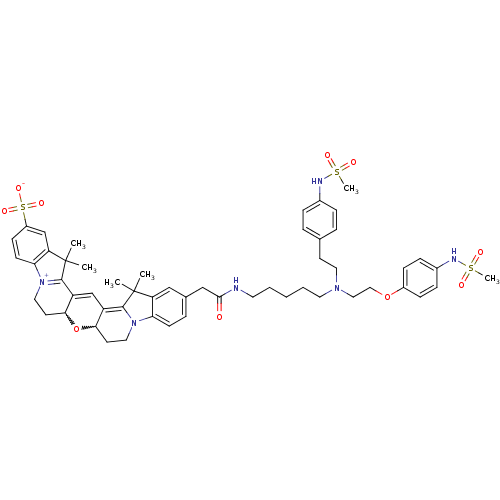
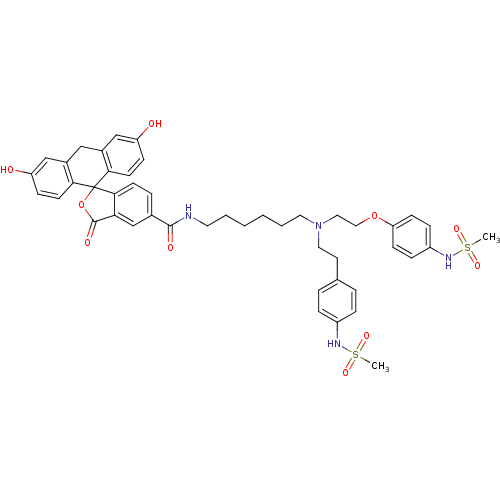
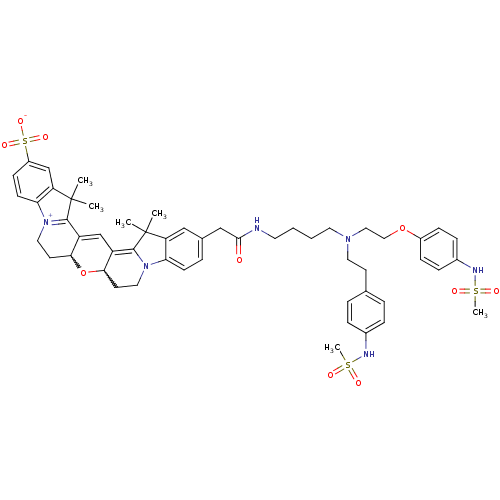
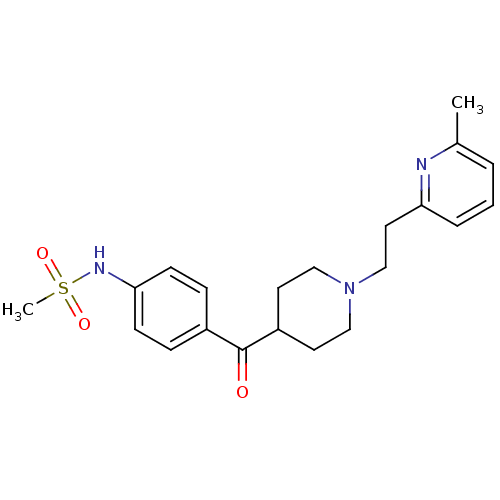
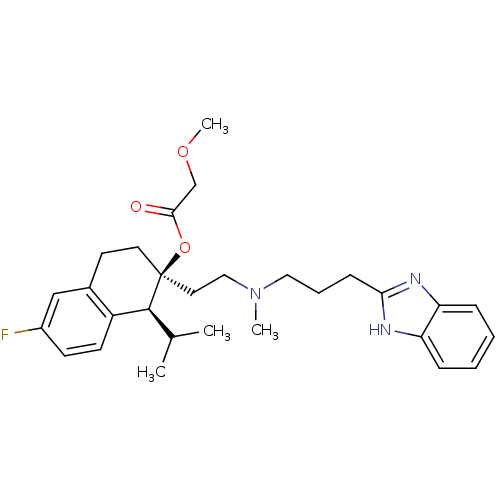
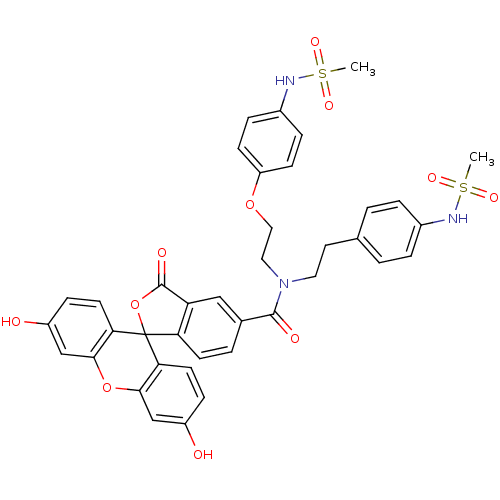
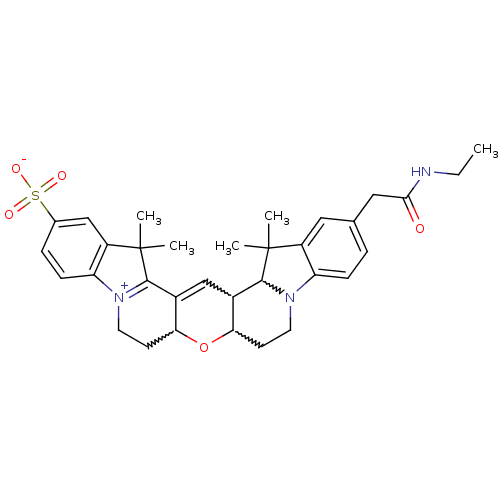
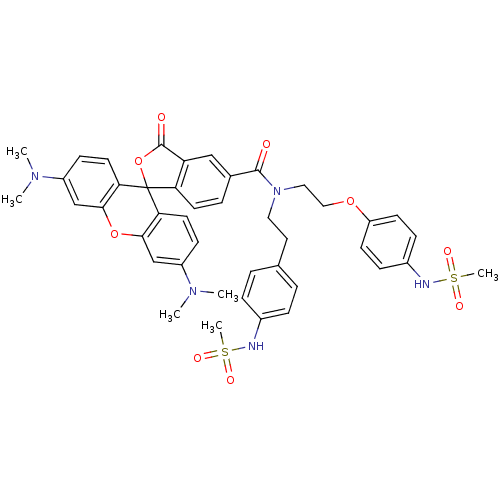
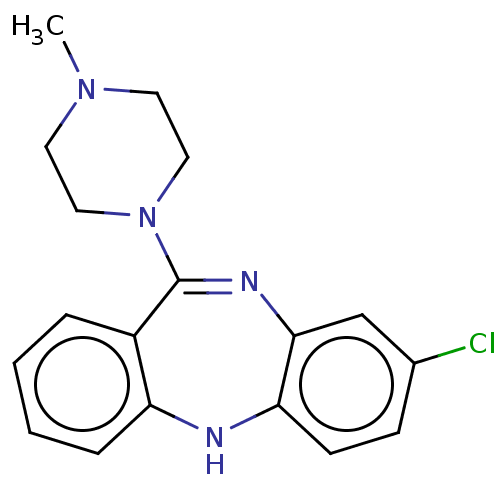
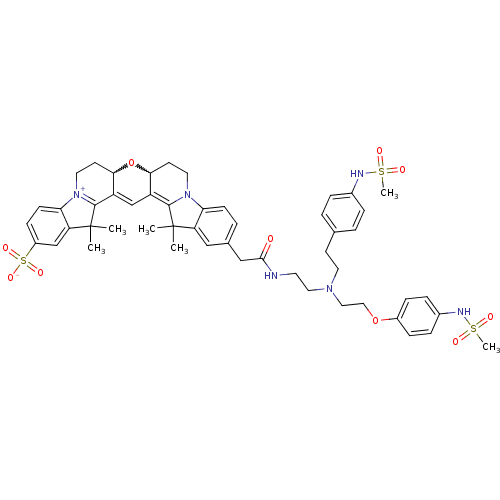
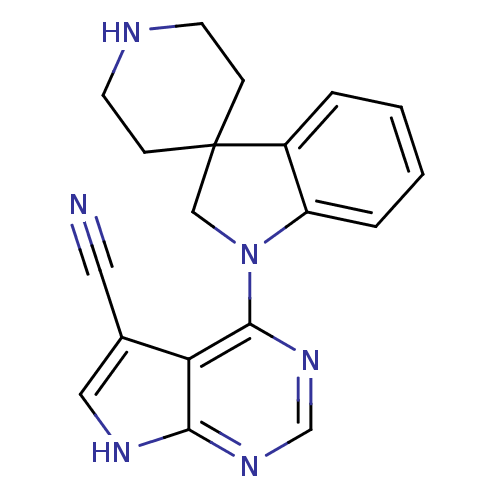
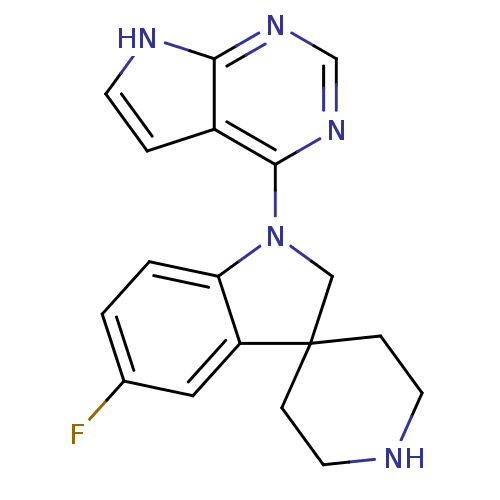
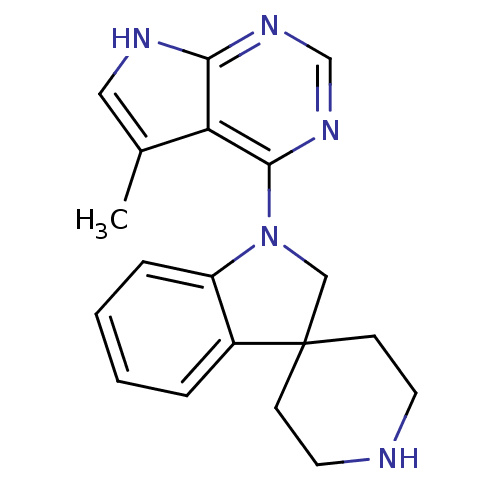
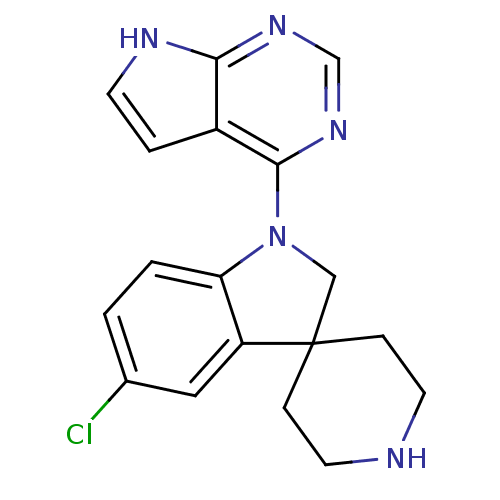

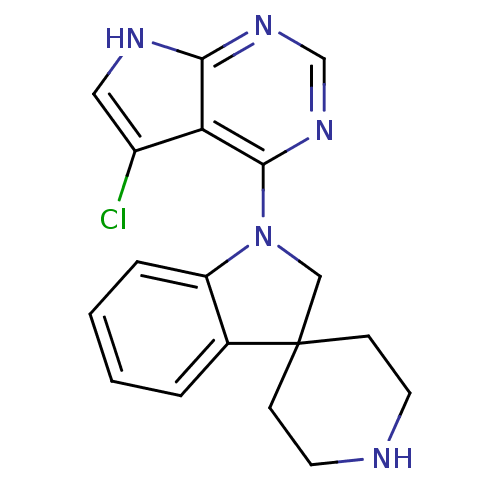
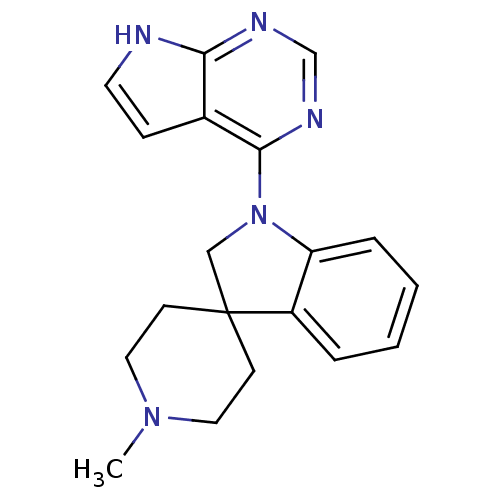

 3D Structure (crystal)
3D Structure (crystal)