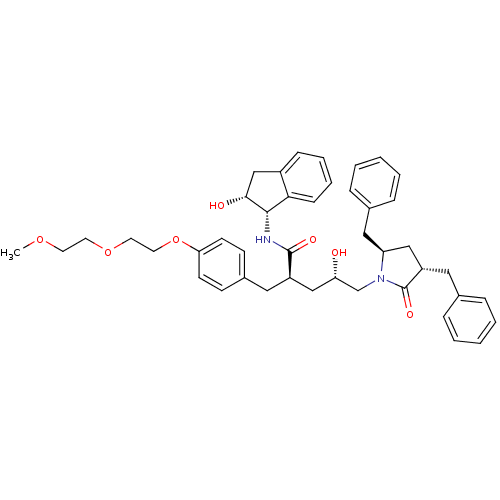
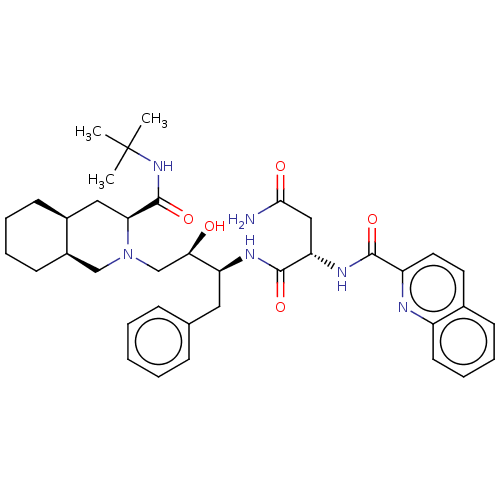
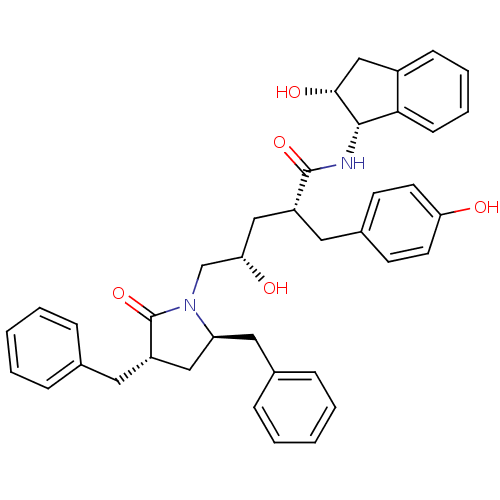
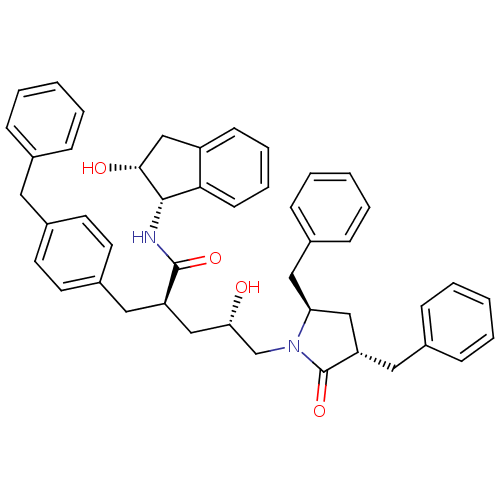
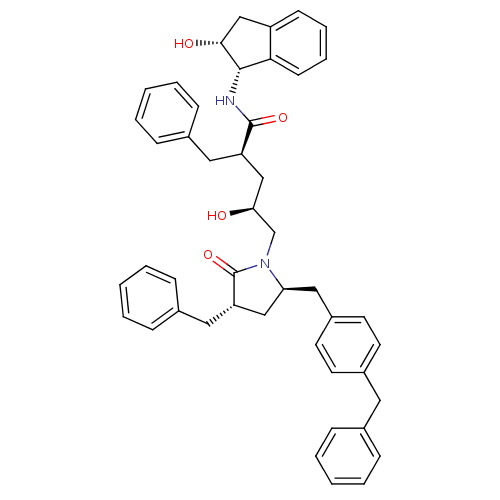
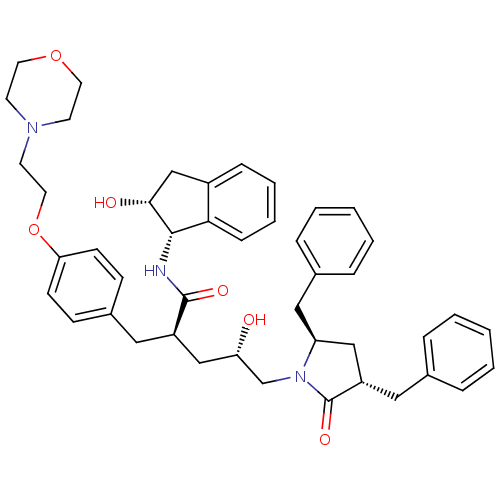
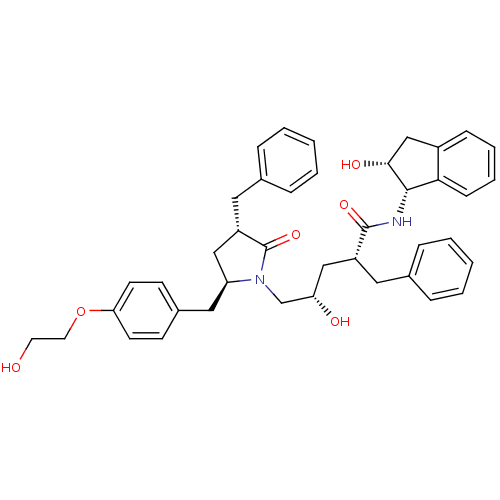
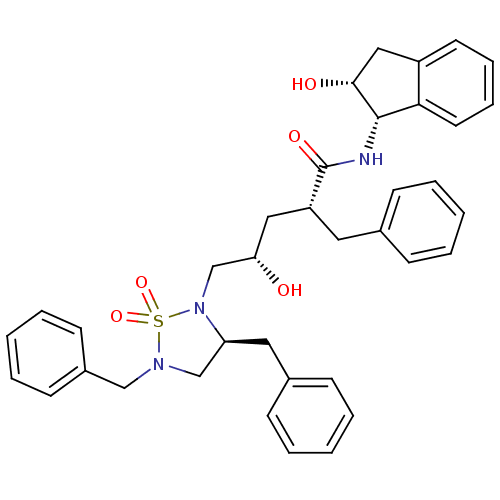
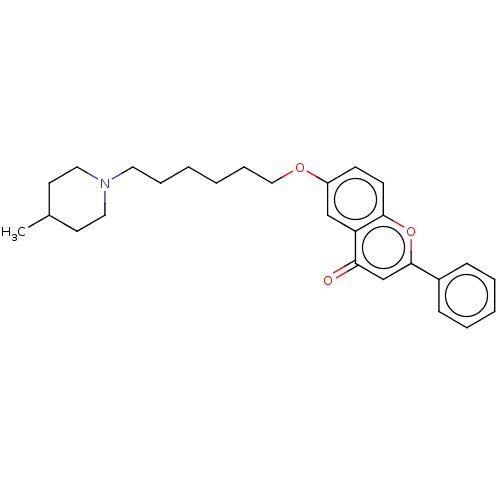
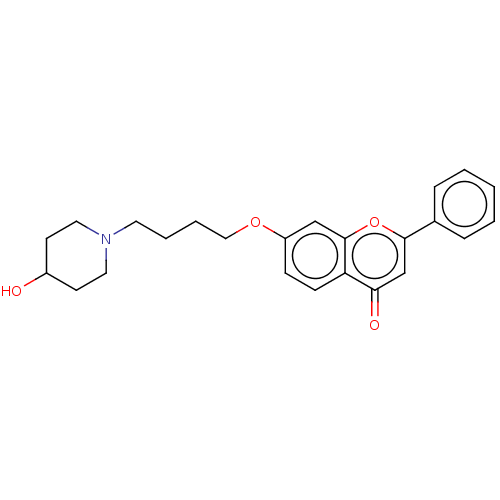
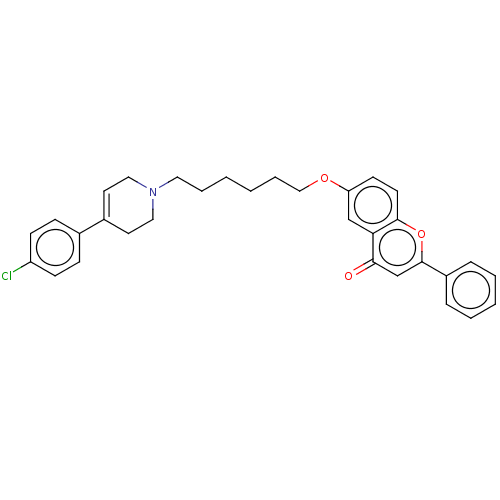
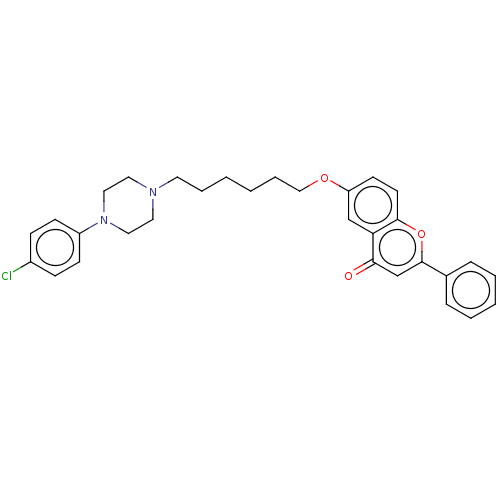
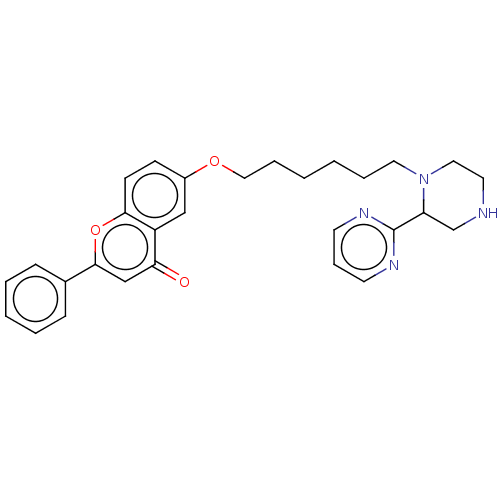
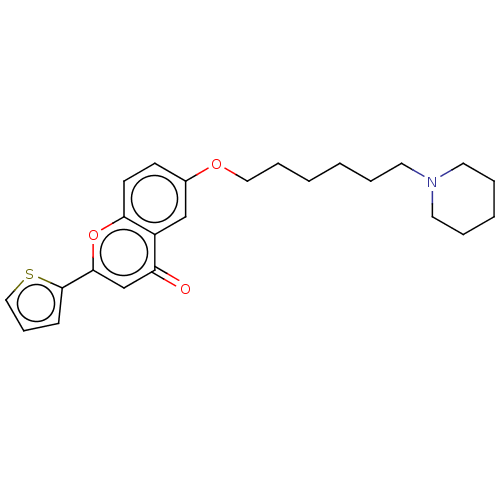
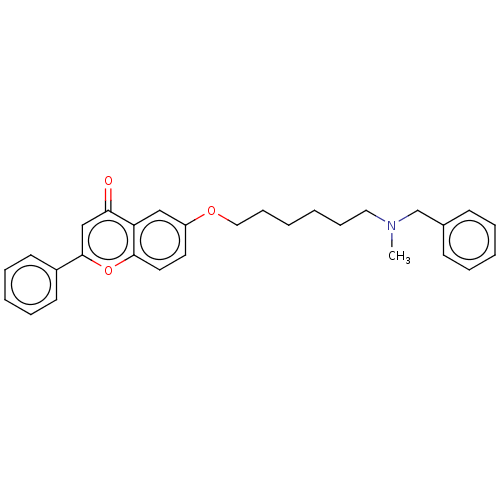
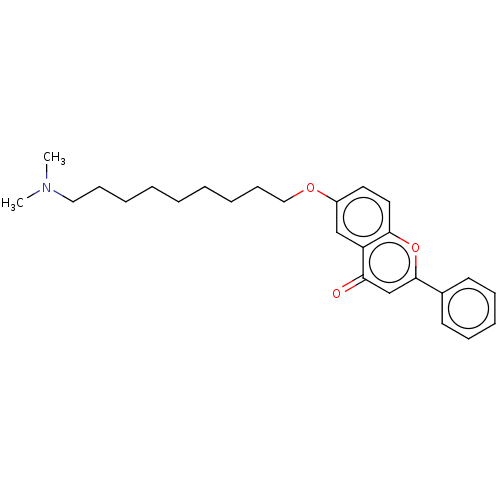
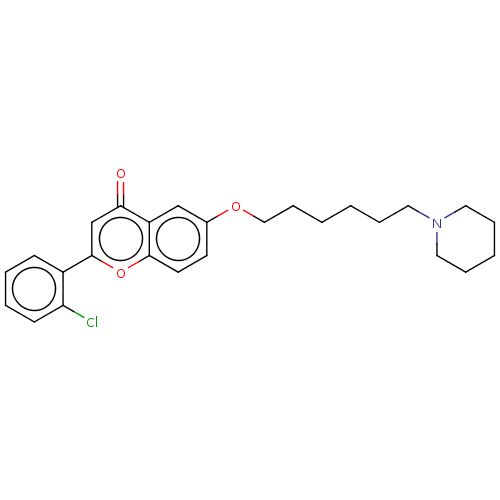
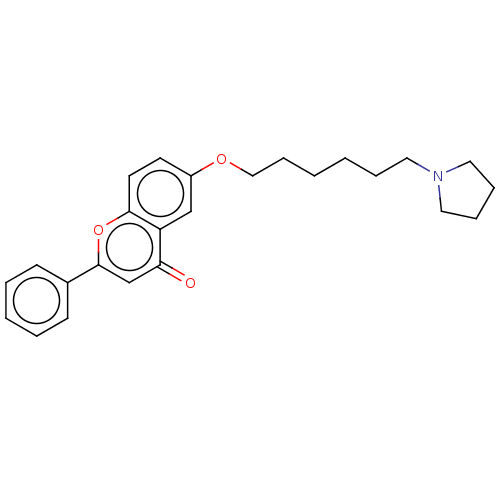
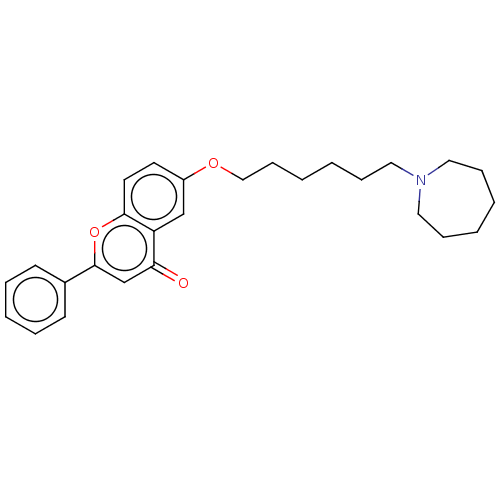
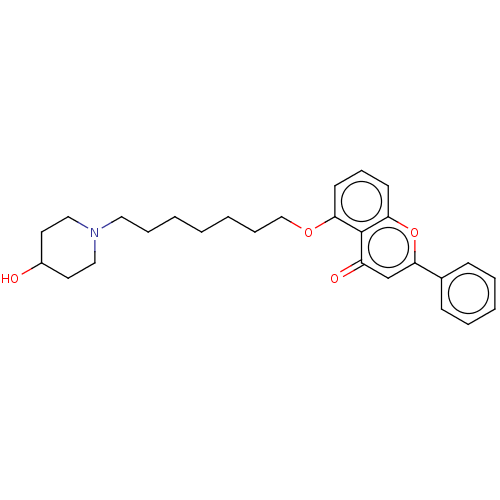
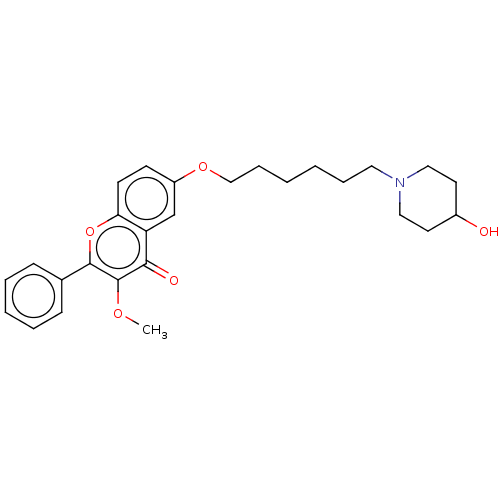
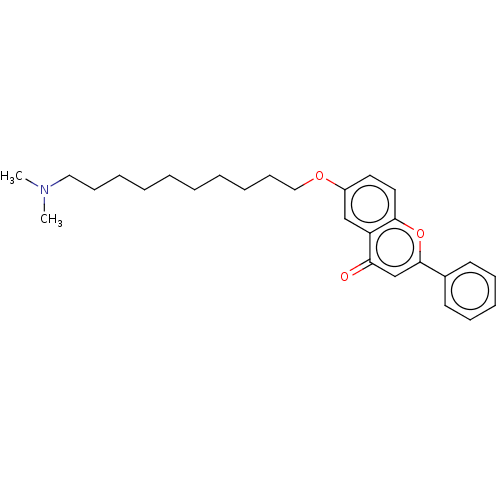
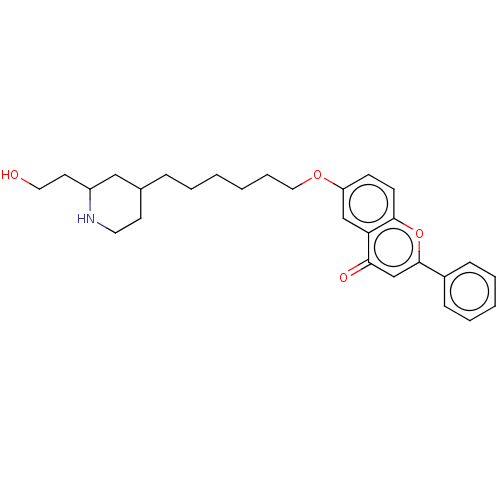
TargetDimer of Gag-Pol polyprotein [501-599,Q508K,L534I,L564I,C568A,C596A](Human immunodeficiency virus type 1)
Glaxosmithkline
Glaxosmithkline
Affinity DataKi: 0.0100nM ΔG°: -62.8kJ/molepH: 6.4 T: 2°CAssay Description:The Ki values were determined using fluorogenic substrate, 2-(aminobenzoyl)-Thr-Ile-Nle-Phe(p-NO2)-Gln-ArgNH2. A standard curve relating changes in f...More data for this Ligand-Target Pair
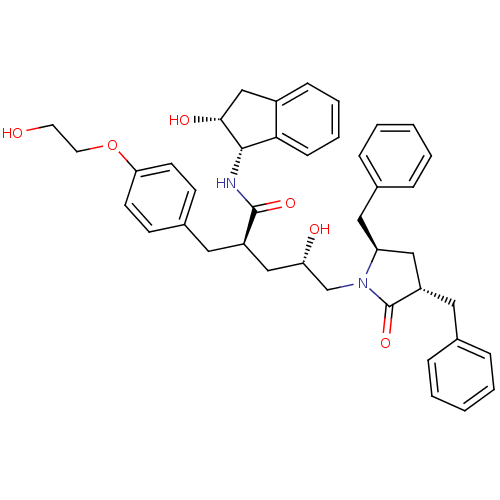
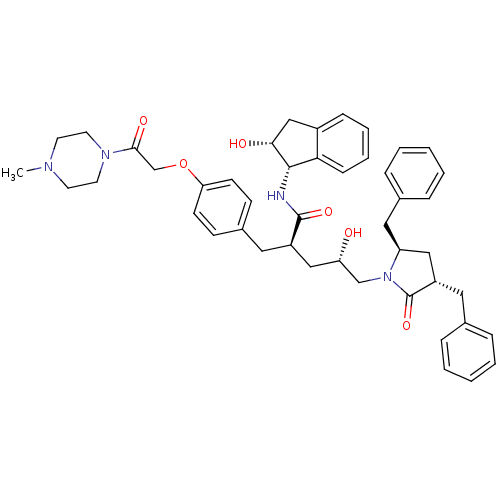
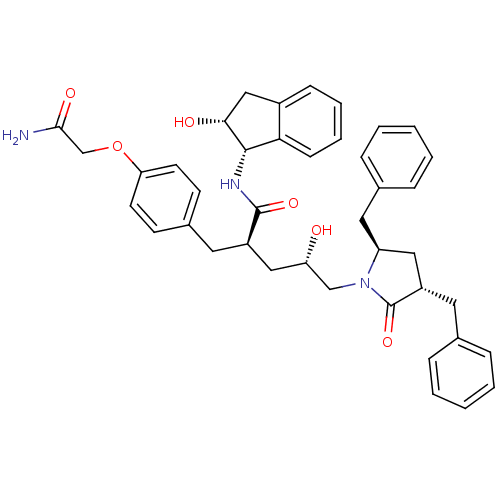
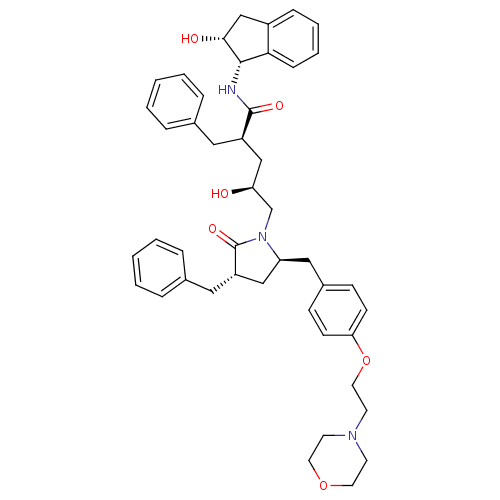
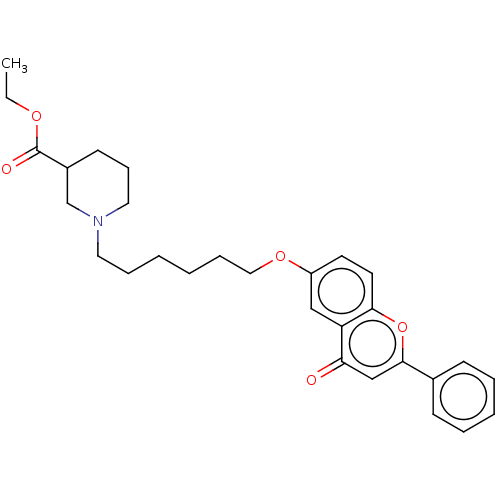
TargetDimer of Gag-Pol polyprotein [501-599,Q508K,L534I,L564I,C568A,C596A](Human immunodeficiency virus type 1)
Glaxosmithkline
Glaxosmithkline
Affinity DataKi: 0.0200nM ΔG°: -61.1kJ/mole IC50: 63nMpH: 6.4 T: 2°CAssay Description:The Ki values were determined using fluorogenic substrate, 2-(aminobenzoyl)-Thr-Ile-Nle-Phe(p-NO2)-Gln-ArgNH2. A standard curve relating changes in f...More data for this Ligand-Target Pair
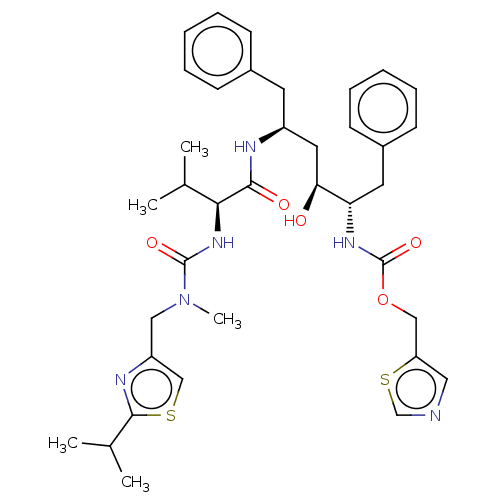
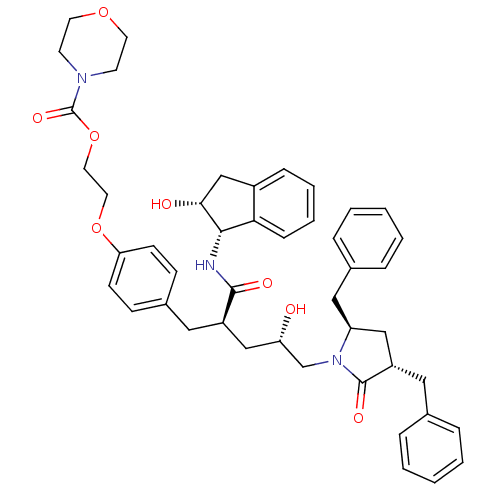
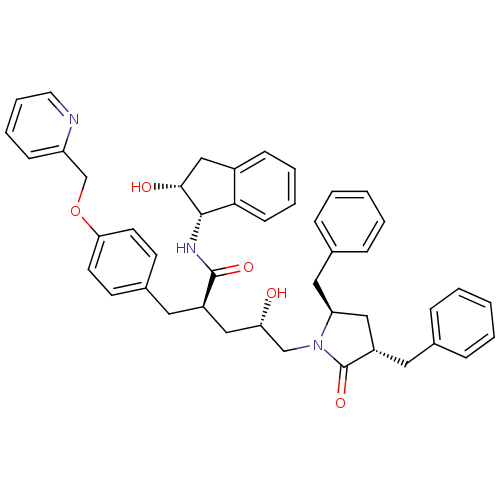
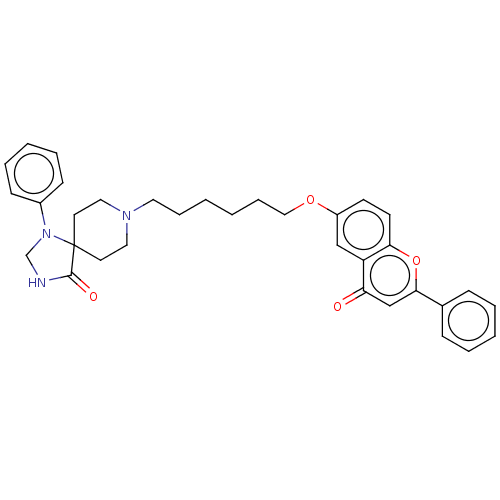
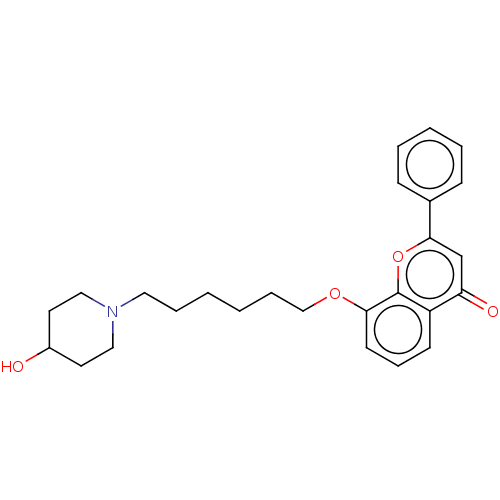
TargetDimer of Gag-Pol polyprotein [501-599,Q508K,L534I,L564I,C568A,C596A](Human immunodeficiency virus type 1)
Glaxosmithkline
Glaxosmithkline
Affinity DataKi: 0.0200nM ΔG°: -61.1kJ/mole IC50: 93nMpH: 6.4 T: 2°CAssay Description:The Ki values were determined using fluorogenic substrate, 2-(aminobenzoyl)-Thr-Ile-Nle-Phe(p-NO2)-Gln-ArgNH2. A standard curve relating changes in f...More data for this Ligand-Target Pair
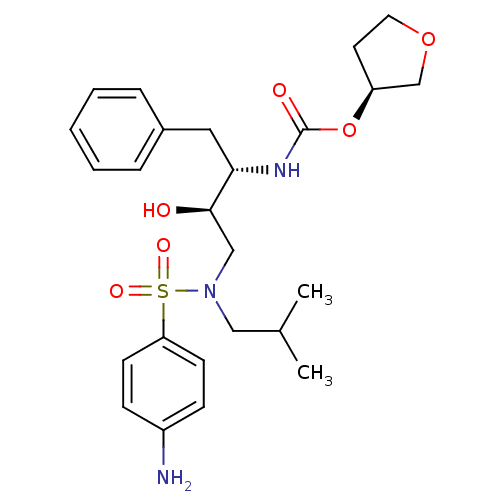
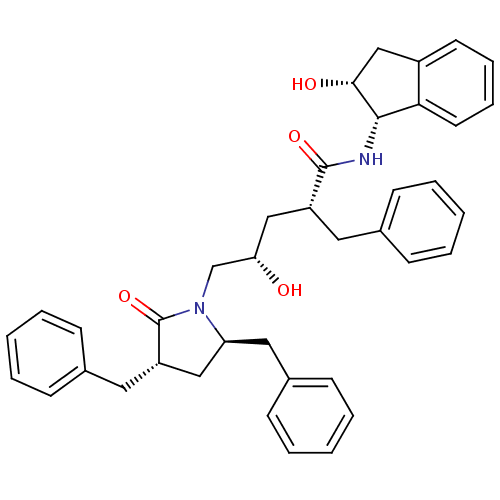
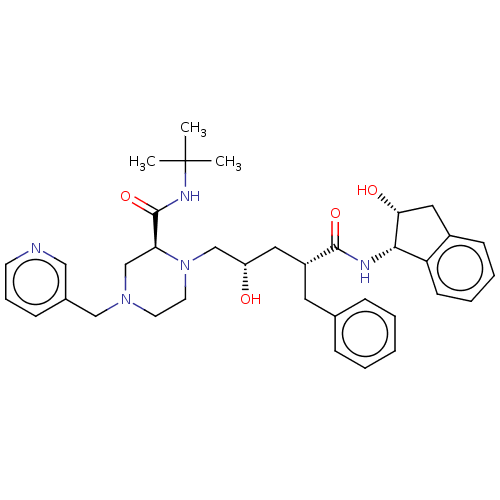
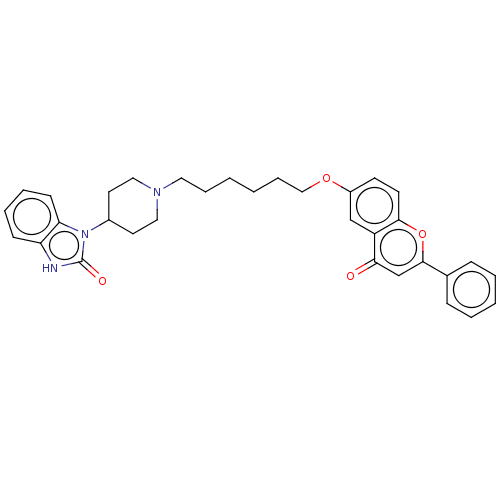
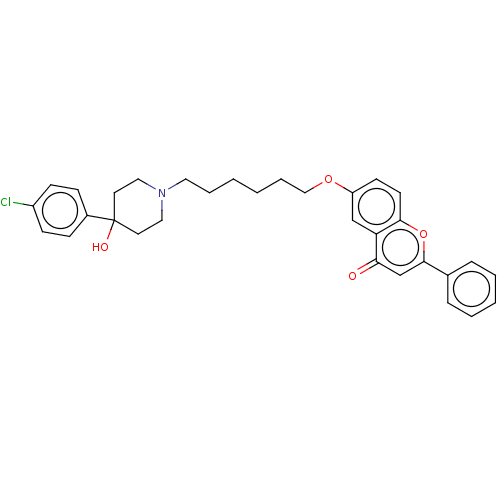
TargetDimer of Gag-Pol polyprotein [501-599,Q508K,L534I,L564I,C568A,C596A](Human immunodeficiency virus type 1)
Glaxosmithkline
Glaxosmithkline
Affinity DataKi: 0.0200nM ΔG°: -61.1kJ/mole IC50: 94nMpH: 6.4 T: 2°CAssay Description:The Ki values were determined using fluorogenic substrate, 2-(aminobenzoyl)-Thr-Ile-Nle-Phe(p-NO2)-Gln-ArgNH2. A standard curve relating changes in f...More data for this Ligand-Target Pair
TargetDimer of Gag-Pol polyprotein [501-599,Q508K,L534I,L564I,C568A,C596A](Human immunodeficiency virus type 1)
Glaxosmithkline
Glaxosmithkline
Affinity DataKi: 0.0200nM ΔG°: -61.1kJ/molepH: 6.4 T: 2°CAssay Description:The Ki values were determined using fluorogenic substrate, 2-(aminobenzoyl)-Thr-Ile-Nle-Phe(p-NO2)-Gln-ArgNH2. A standard curve relating changes in f...More data for this Ligand-Target Pair
TargetDimer of Gag-Pol polyprotein [501-599,Q508K,L534I,L564I,C568A,C596A](Human immunodeficiency virus type 1)
Glaxosmithkline
Glaxosmithkline
Affinity DataKi: 0.0400nM ΔG°: -59.3kJ/mole IC50: 150nMpH: 6.4 T: 2°CAssay Description:The Ki values were determined using fluorogenic substrate, 2-(aminobenzoyl)-Thr-Ile-Nle-Phe(p-NO2)-Gln-ArgNH2. A standard curve relating changes in f...More data for this Ligand-Target Pair
TargetDimer of Gag-Pol polyprotein [501-599,Q508K,L534I,L564I,C568A,C596A](Human immunodeficiency virus type 1)
Glaxosmithkline
Glaxosmithkline
Affinity DataKi: 0.0400nM ΔG°: -59.3kJ/molepH: 6.4 T: 2°CAssay Description:The Ki values were determined using fluorogenic substrate, 2-(aminobenzoyl)-Thr-Ile-Nle-Phe(p-NO2)-Gln-ArgNH2. A standard curve relating changes in f...More data for this Ligand-Target Pair
TargetDimer of Gag-Pol polyprotein [501-599,Q508K,L534I,L564I,C568A,C596A](Human immunodeficiency virus type 1)
Glaxosmithkline
Glaxosmithkline
Affinity DataKi: 0.0400nM ΔG°: -59.3kJ/molepH: 6.4 T: 2°CAssay Description:The Ki values were determined using fluorogenic substrate, 2-(aminobenzoyl)-Thr-Ile-Nle-Phe(p-NO2)-Gln-ArgNH2. A standard curve relating changes in f...More data for this Ligand-Target Pair
TargetDimer of Gag-Pol polyprotein [501-599,Q508K,L534I,L564I,C568A,C596A](Human immunodeficiency virus type 1)
Glaxosmithkline
Glaxosmithkline
Affinity DataKi: 0.0500nM ΔG°: -58.8kJ/mole IC50: 130nMpH: 6.4 T: 2°CAssay Description:The Ki values were determined using fluorogenic substrate, 2-(aminobenzoyl)-Thr-Ile-Nle-Phe(p-NO2)-Gln-ArgNH2. A standard curve relating changes in f...More data for this Ligand-Target Pair
TargetDimer of Gag-Pol polyprotein [501-599,Q508K,L534I,L564I,C568A,C596A](Human immunodeficiency virus type 1)
Glaxosmithkline
Glaxosmithkline
Affinity DataKi: 0.0500nM ΔG°: -58.8kJ/mole IC50: 240nMpH: 6.4 T: 2°CAssay Description:The Ki values were determined using fluorogenic substrate, 2-(aminobenzoyl)-Thr-Ile-Nle-Phe(p-NO2)-Gln-ArgNH2. A standard curve relating changes in f...More data for this Ligand-Target Pair
TargetDimer of Gag-Pol polyprotein [501-599,Q508K,L534I,L564I,C568A,C596A](Human immunodeficiency virus type 1)
Glaxosmithkline
Glaxosmithkline
Affinity DataKi: 0.0500nM ΔG°: -58.8kJ/mole IC50: 720nMpH: 6.4 T: 2°CAssay Description:The Ki values were determined using fluorogenic substrate, 2-(aminobenzoyl)-Thr-Ile-Nle-Phe(p-NO2)-Gln-ArgNH2. A standard curve relating changes in f...More data for this Ligand-Target Pair
TargetDimer of Gag-Pol polyprotein [501-599,Q508K,L534I,L564I,C568A,C596A](Human immunodeficiency virus type 1)
Glaxosmithkline
Glaxosmithkline
Affinity DataKi: 0.0500nM ΔG°: -58.8kJ/molepH: 6.4 T: 2°CAssay Description:The Ki values were determined using fluorogenic substrate, 2-(aminobenzoyl)-Thr-Ile-Nle-Phe(p-NO2)-Gln-ArgNH2. A standard curve relating changes in f...More data for this Ligand-Target Pair
TargetDimer of Gag-Pol polyprotein [501-599,Q508K,L534I,L564I,C568A,C596A](Human immunodeficiency virus type 1)
Glaxosmithkline
Glaxosmithkline
Affinity DataKi: 0.0600nM ΔG°: -58.3kJ/mole IC50: 320nMpH: 6.4 T: 2°CAssay Description:The Ki values were determined using fluorogenic substrate, 2-(aminobenzoyl)-Thr-Ile-Nle-Phe(p-NO2)-Gln-ArgNH2. A standard curve relating changes in f...More data for this Ligand-Target Pair
TargetDimer of Gag-Pol polyprotein [501-599,Q508K,L534I,L564I,C568A,C596A](Human immunodeficiency virus type 1)
Glaxosmithkline
Glaxosmithkline
Affinity DataKi: 0.0700nM ΔG°: -58.0kJ/mole IC50: 360nMpH: 6.4 T: 2°CAssay Description:The Ki values were determined using fluorogenic substrate, 2-(aminobenzoyl)-Thr-Ile-Nle-Phe(p-NO2)-Gln-ArgNH2. A standard curve relating changes in f...More data for this Ligand-Target Pair
TargetDimer of Gag-Pol polyprotein [501-599,Q508K,L534I,L564I,C568A,C596A](Human immunodeficiency virus type 1)
Glaxosmithkline
Glaxosmithkline
Affinity DataKi: 0.0700nM ΔG°: -58.0kJ/mole IC50: 380nMpH: 6.4 T: 2°CAssay Description:The Ki values were determined using fluorogenic substrate, 2-(aminobenzoyl)-Thr-Ile-Nle-Phe(p-NO2)-Gln-ArgNH2. A standard curve relating changes in f...More data for this Ligand-Target Pair
TargetDimer of Gag-Pol polyprotein [501-599,Q508K,L534I,L564I,C568A,C596A](Human immunodeficiency virus type 1)
Glaxosmithkline
Glaxosmithkline
Affinity DataKi: 0.0700nM ΔG°: -58.0kJ/molepH: 6.4 T: 2°CAssay Description:The Ki values were determined using fluorogenic substrate, 2-(aminobenzoyl)-Thr-Ile-Nle-Phe(p-NO2)-Gln-ArgNH2. A standard curve relating changes in f...More data for this Ligand-Target Pair
TargetDimer of Gag-Pol polyprotein [501-599,Q508K,L534I,L564I,C568A,C596A](Human immunodeficiency virus type 1)
Glaxosmithkline
Glaxosmithkline
Affinity DataKi: 0.100nM ΔG°: -57.1kJ/mole IC50: 200nMpH: 6.4 T: 2°CAssay Description:The Ki values were determined using fluorogenic substrate, 2-(aminobenzoyl)-Thr-Ile-Nle-Phe(p-NO2)-Gln-ArgNH2. A standard curve relating changes in f...More data for this Ligand-Target Pair
TargetDimer of Gag-Pol polyprotein [501-599,Q508K,L534I,L564I,C568A,C596A](Human immunodeficiency virus type 1)
Glaxosmithkline
Glaxosmithkline
Affinity DataKi: 0.110nM ΔG°: -56.8kJ/mole IC50: 920nMpH: 6.4 T: 2°CAssay Description:The Ki values were determined using fluorogenic substrate, 2-(aminobenzoyl)-Thr-Ile-Nle-Phe(p-NO2)-Gln-ArgNH2. A standard curve relating changes in f...More data for this Ligand-Target Pair
TargetDimer of Gag-Pol polyprotein [501-599,Q508K,L534I,L564I,C568A,C596A](Human immunodeficiency virus type 1)
Glaxosmithkline
Glaxosmithkline
Affinity DataKi: 0.130nM ΔG°: -56.4kJ/mole IC50: 200nMpH: 6.4 T: 2°CAssay Description:The Ki values were determined using fluorogenic substrate, 2-(aminobenzoyl)-Thr-Ile-Nle-Phe(p-NO2)-Gln-ArgNH2. A standard curve relating changes in f...More data for this Ligand-Target Pair
TargetDimer of Gag-Pol polyprotein [501-599,Q508K,L534I,L564I,C568A,C596A](Human immunodeficiency virus type 1)
Glaxosmithkline
Glaxosmithkline
Affinity DataKi: 0.130nM ΔG°: -56.4kJ/mole IC50: 550nMpH: 6.4 T: 2°CAssay Description:The Ki values were determined using fluorogenic substrate, 2-(aminobenzoyl)-Thr-Ile-Nle-Phe(p-NO2)-Gln-ArgNH2. A standard curve relating changes in f...More data for this Ligand-Target Pair
TargetDimer of Gag-Pol polyprotein [501-599,Q508K,L534I,L564I,C568A,C596A](Human immunodeficiency virus type 1)
Glaxosmithkline
Glaxosmithkline
Affinity DataKi: 0.140nM ΔG°: -56.2kJ/mole IC50: 83nMpH: 6.4 T: 2°CAssay Description:The Ki values were determined using fluorogenic substrate, 2-(aminobenzoyl)-Thr-Ile-Nle-Phe(p-NO2)-Gln-ArgNH2. A standard curve relating changes in f...More data for this Ligand-Target Pair
TargetDimer of Gag-Pol polyprotein [501-599,Q508K,L534I,L564I,C568A,C596A](Human immunodeficiency virus type 1)
Glaxosmithkline
Glaxosmithkline
Affinity DataKi: 0.300nM ΔG°: -54.4kJ/mole IC50: 480nMpH: 6.4 T: 2°CAssay Description:The Ki values were determined using fluorogenic substrate, 2-(aminobenzoyl)-Thr-Ile-Nle-Phe(p-NO2)-Gln-ArgNH2. A standard curve relating changes in f...More data for this Ligand-Target Pair
TargetDimer of Gag-Pol polyprotein [501-599,Q508K,L534I,L564I,C568A,C596A](Human immunodeficiency virus type 1)
Glaxosmithkline
Glaxosmithkline
Affinity DataKi: >0.5nM ΔG°: >-53.1kJ/molepH: 6.4 T: 2°CAssay Description:The Ki values were determined using fluorogenic substrate, 2-(aminobenzoyl)-Thr-Ile-Nle-Phe(p-NO2)-Gln-ArgNH2. A standard curve relating changes in f...More data for this Ligand-Target Pair
TargetDimer of Gag-Pol polyprotein [501-599,Q508K,L534I,L564I,C568A,C596A](Human immunodeficiency virus type 1)
Glaxosmithkline
Glaxosmithkline
Affinity DataKi: 0.800nM ΔG°: -51.9kJ/molepH: 6.4 T: 2°CAssay Description:The Ki values were determined using fluorogenic substrate, 2-(aminobenzoyl)-Thr-Ile-Nle-Phe(p-NO2)-Gln-ArgNH2. A standard curve relating changes in f...More data for this Ligand-Target Pair
TargetDimer of Gag-Pol polyprotein [501-599,Q508K,L534I,L564I,C568A,C596A](Human immunodeficiency virus type 1)
Glaxosmithkline
Glaxosmithkline
Affinity DataKi: 1.20nM ΔG°: -50.9kJ/molepH: 6.4 T: 2°CAssay Description:The Ki values were determined using fluorogenic substrate, 2-(aminobenzoyl)-Thr-Ile-Nle-Phe(p-NO2)-Gln-ArgNH2. A standard curve relating changes in f...More data for this Ligand-Target Pair
TargetDimer of Gag-Pol polyprotein [501-599,Q508K,L534I,L564I,C568A,C596A](Human immunodeficiency virus type 1)
Glaxosmithkline
Glaxosmithkline
Affinity DataKi: 1.30nM ΔG°: -50.7kJ/mole IC50: 960nMpH: 6.4 T: 2°CAssay Description:The Ki values were determined using fluorogenic substrate, 2-(aminobenzoyl)-Thr-Ile-Nle-Phe(p-NO2)-Gln-ArgNH2. A standard curve relating changes in f...More data for this Ligand-Target Pair
TargetDimer of Gag-Pol polyprotein [501-599,Q508K,L534I,L564I,C568A,C596A](Human immunodeficiency virus type 1)
Glaxosmithkline
Glaxosmithkline
Affinity DataKi: 16nM ΔG°: -44.5kJ/molepH: 6.4 T: 2°CAssay Description:The Ki values were determined using fluorogenic substrate, 2-(aminobenzoyl)-Thr-Ile-Nle-Phe(p-NO2)-Gln-ArgNH2. A standard curve relating changes in f...More data for this Ligand-Target Pair
Affinity DataKi: 180nMAssay Description:Affinity towards dopamine receptor D2 site was determined by competition studies using [3H]-sulpiride in rat striatum.More data for this Ligand-Target Pair
Affinity DataKi: 220nMAssay Description:Affinity towards dopamine receptor D2 site was determined by competition studies using [3H]-sulpiride in rat striatum.More data for this Ligand-Target Pair
TargetDimer of Gag-Pol polyprotein [501-599,Q508K,L534I,L564I,C568A,C596A](Human immunodeficiency virus type 1)
Glaxosmithkline
Glaxosmithkline
Affinity DataKi: 330nM ΔG°: -37.0kJ/molepH: 6.4 T: 2°CAssay Description:The Ki values were determined using fluorogenic substrate, 2-(aminobenzoyl)-Thr-Ile-Nle-Phe(p-NO2)-Gln-ArgNH2. A standard curve relating changes in f...More data for this Ligand-Target Pair
Affinity DataKi: 570nMAssay Description:Affinity towards dopamine receptor D2 was determined by competition studies using [3H]-sulpiride in rat striatum.More data for this Ligand-Target Pair
Affinity DataKi: 700nMAssay Description:Affinity towards dopamine receptor D2 site was determined by competition studies using [3H]-sulpiride in rat striatum.More data for this Ligand-Target Pair
Affinity DataKi: 940nMAssay Description:Affinity towards dopamine receptor D2 was determined by competition studies using [3H]-sulpiride in rat striatum.More data for this Ligand-Target Pair
Affinity DataKi: 990nMAssay Description:Affinity towards dopamine receptor D2 site was determined by competition studies using [3H]-sulpiride in rat striatum.More data for this Ligand-Target Pair
Affinity DataKi: 1.00E+3nMAssay Description:Affinity towards dopamine receptor D2 was determined by competition studies using [3H]-sulpiride in rat striatum.More data for this Ligand-Target Pair
Affinity DataKi: 1.30E+3nMAssay Description:Affinity towards dopamine receptor D2 site was determined by competition studies using [3H]-sulpiride in rat striatum.More data for this Ligand-Target Pair
Affinity DataKi: 1.30E+3nMAssay Description:Affinity towards dopamine receptor D2 site was determined by competition studies using [3H]-sulpiride in rat striatum.More data for this Ligand-Target Pair
Affinity DataKi: 1.30E+3nMAssay Description:Affinity towards dopamine receptor D2 was determined by competition studies using [3H]-sulpiride in rat striatum.More data for this Ligand-Target Pair
Affinity DataKi: 1.40E+3nMAssay Description:Affinity towards dopamine receptor D2 was determined by competition studies using [3H]-sulpiride in rat striatum.More data for this Ligand-Target Pair
Affinity DataKi: 1.80E+3nMAssay Description:Affinity towards dopamine receptor D2 site was determined by competition studies using [3H]-sulpiride in rat striatum.More data for this Ligand-Target Pair
Affinity DataKi: 1.80E+3nMAssay Description:Affinity towards dopamine receptor D2 was determined by competition studies using [3H]-sulpiride in rat striatum.More data for this Ligand-Target Pair
Affinity DataKi: 2.20E+3nMAssay Description:Affinity towards dopamine receptor D2 was determined by competition studies using [3H]-sulpiride in rat striatum.More data for this Ligand-Target Pair
Affinity DataKi: 2.20E+3nMAssay Description:Affinity towards dopamine receptor D2 was determined by competition studies using [3H]-sulpiride in rat striatum.More data for this Ligand-Target Pair
Affinity DataKi: 2.30E+3nMAssay Description:Affinity towards dopamine receptor D2 was determined by competition studies using [3H]-sulpiride in rat striatum.More data for this Ligand-Target Pair
Affinity DataKi: 2.30E+3nMAssay Description:Affinity towards dopamine receptor D2 site was determined by competition studies using [3H]-sulpiride in rat striatum.More data for this Ligand-Target Pair
Affinity DataKi: 2.30E+3nMAssay Description:Affinity towards dopamine receptor D2 was determined by competition studies using [3H]-sulpiride in rat striatum.More data for this Ligand-Target Pair
Affinity DataKi: 2.40E+3nMAssay Description:Affinity towards dopamine receptor D2 was determined by competition studies using [3H]-sulpiride in rat striatum.More data for this Ligand-Target Pair
Affinity DataKi: 2.60E+3nMAssay Description:Affinity towards dopamine receptor D2 was determined by competition studies using [3H]-sulpiride in rat striatum.More data for this Ligand-Target Pair
Affinity DataKi: 2.70E+3nMAssay Description:Affinity towards dopamine receptor D2 was determined by competition studies using [3H]-sulpiride in rat striatum.More data for this Ligand-Target Pair
Affinity DataKi: 2.70E+3nMAssay Description:Affinity towards dopamine receptor D2 was determined by competition studies using [3H]-sulpiride in rat striatum.More data for this Ligand-Target Pair

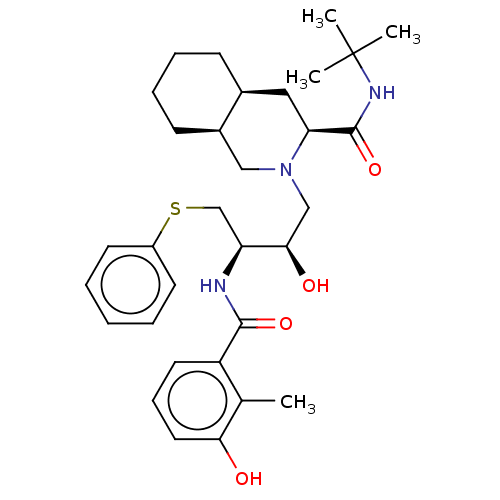
 3D Structure (crystal)
3D Structure (crystal)