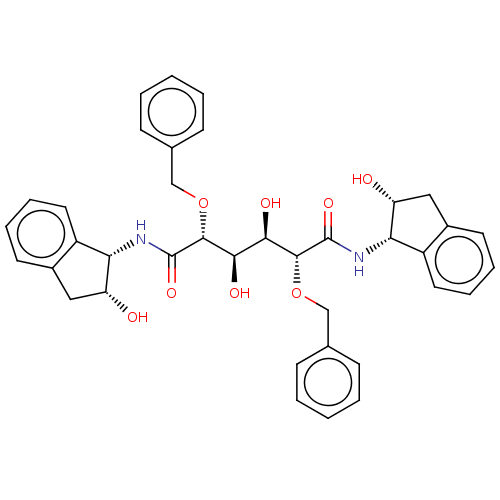
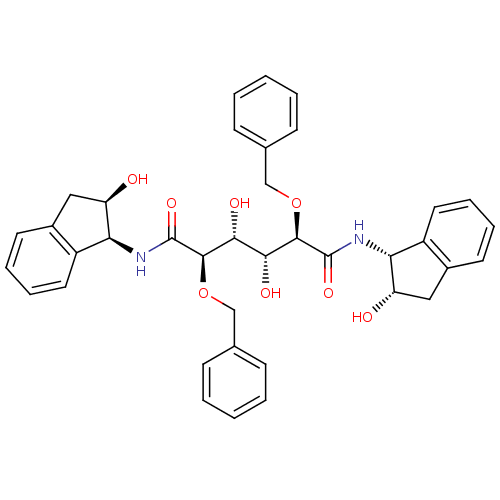
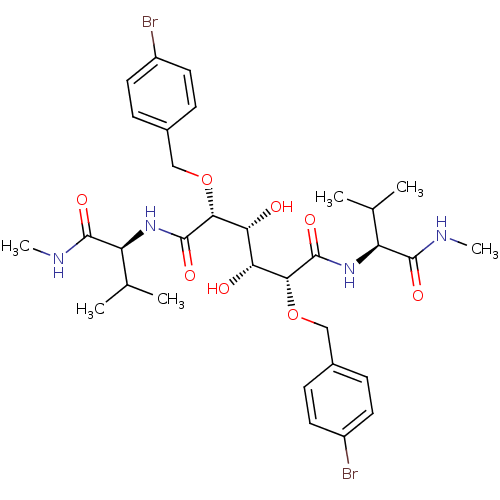
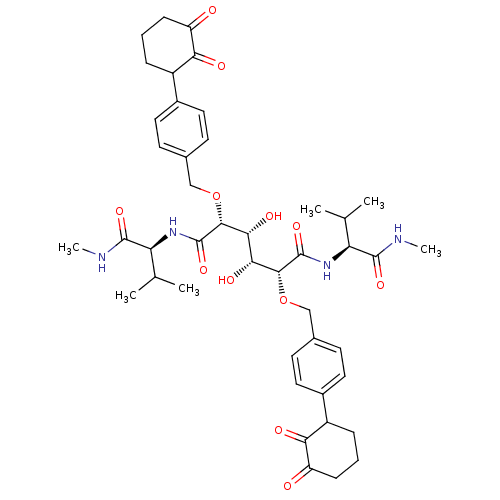
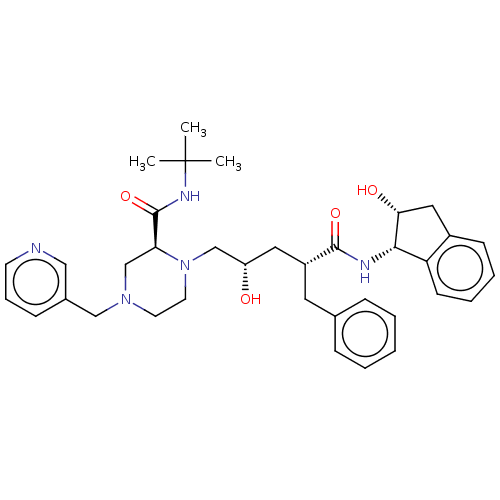
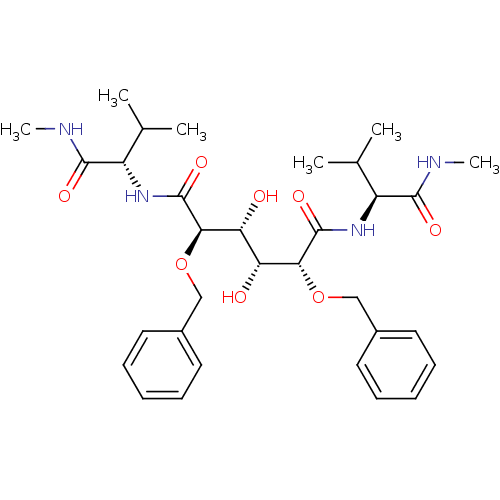
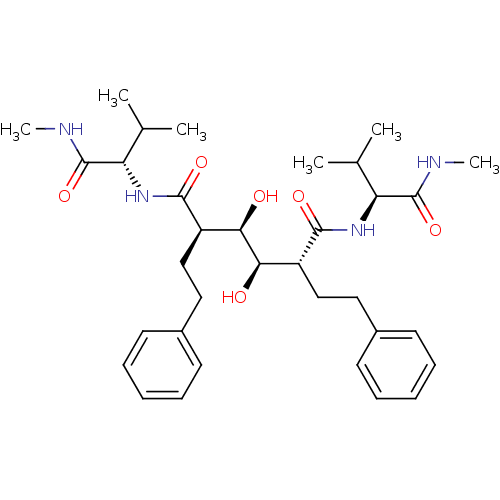
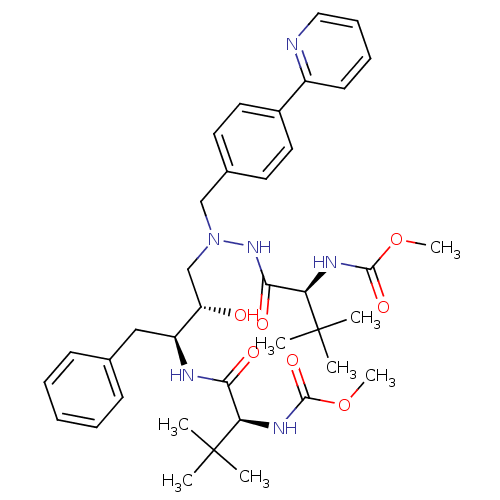
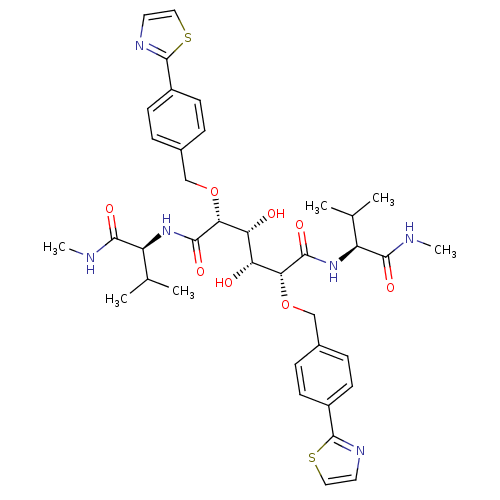
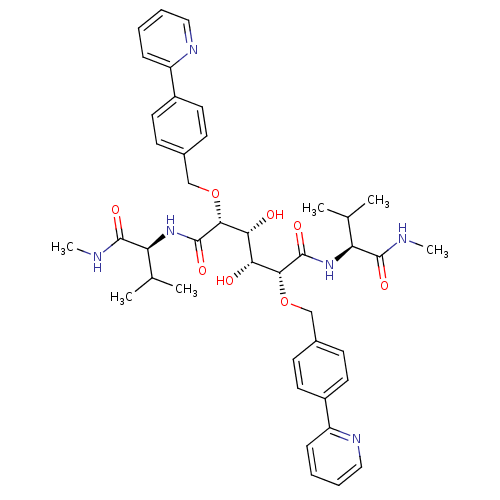
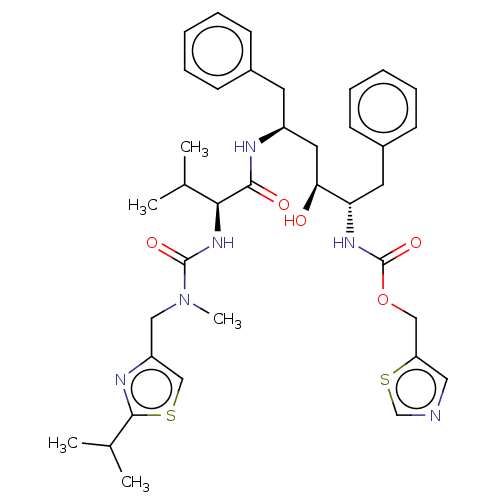
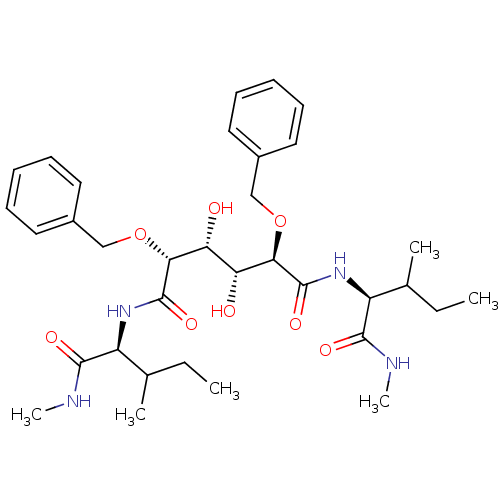
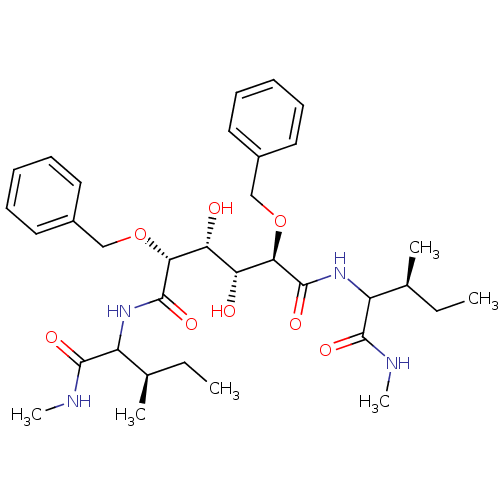
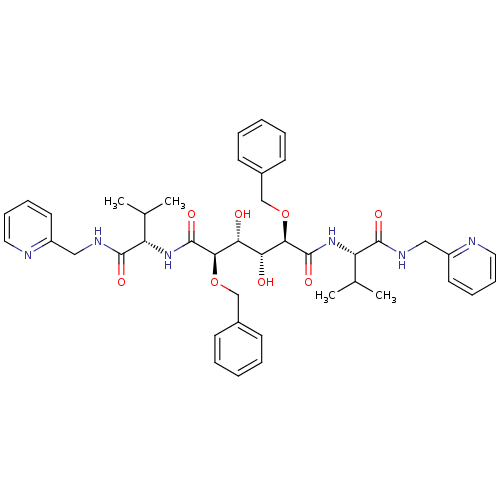
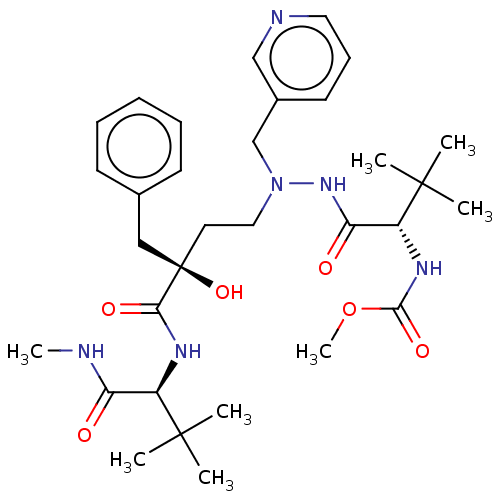
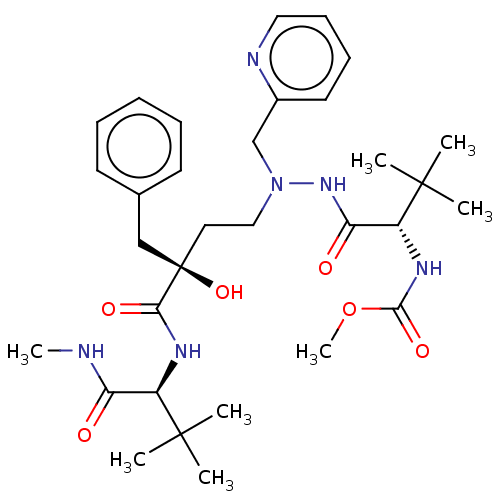
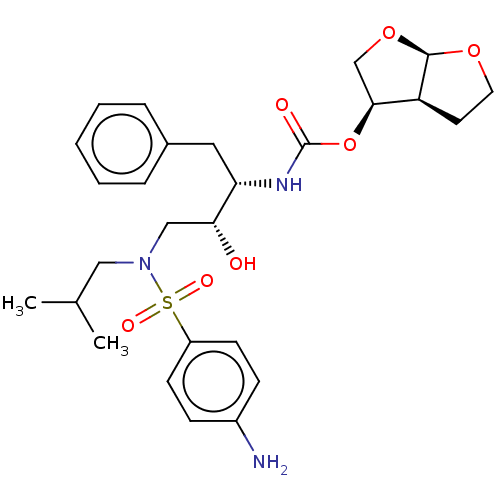
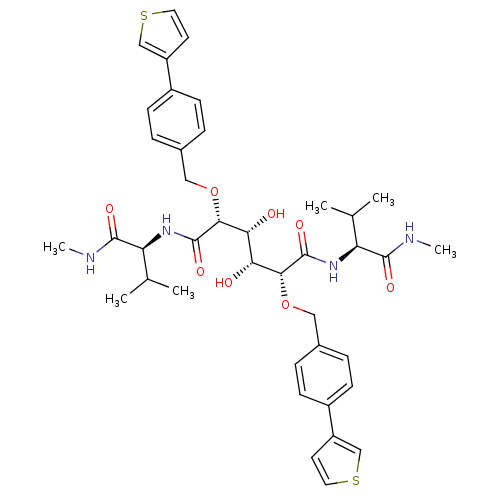
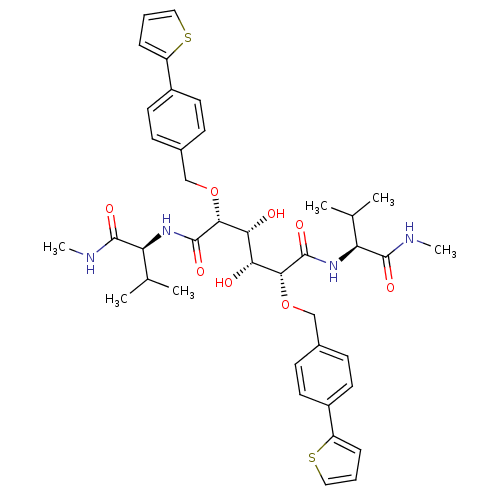
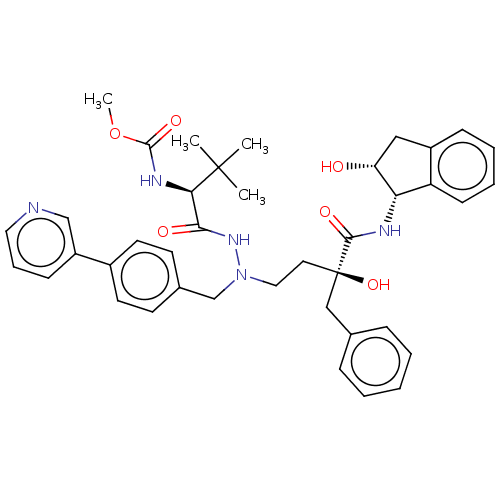
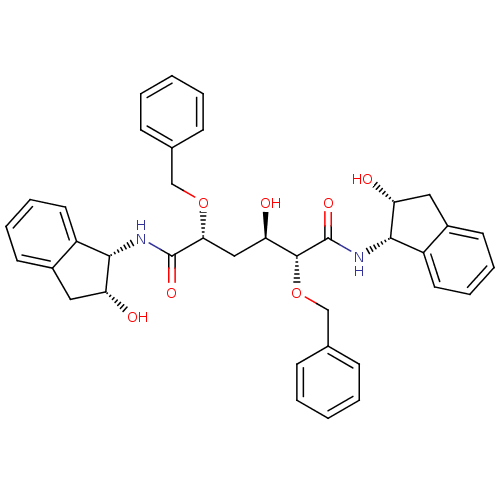
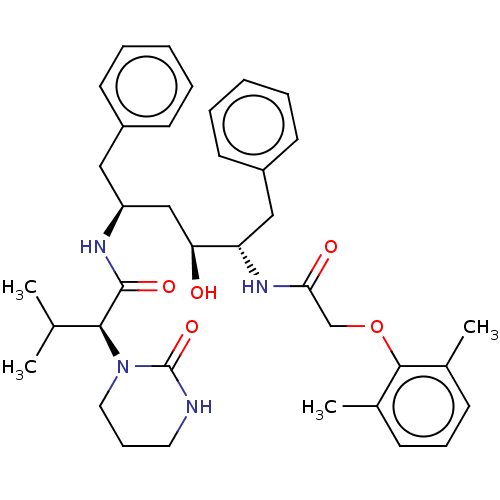
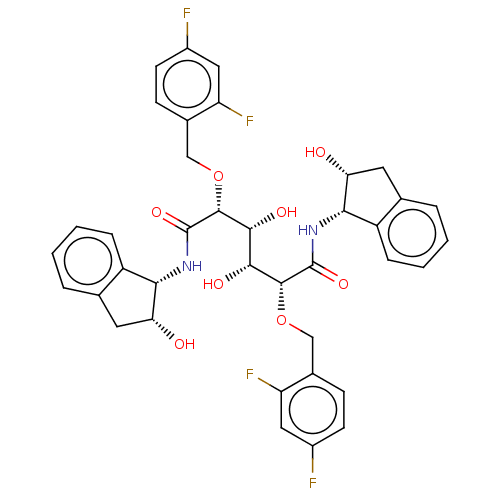
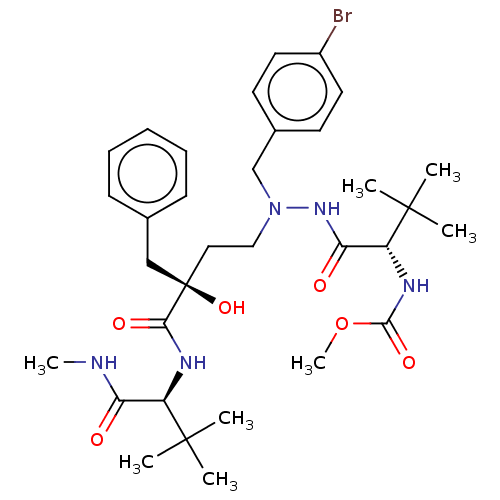
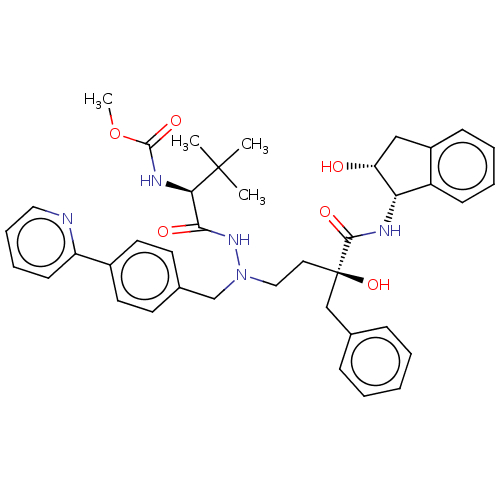
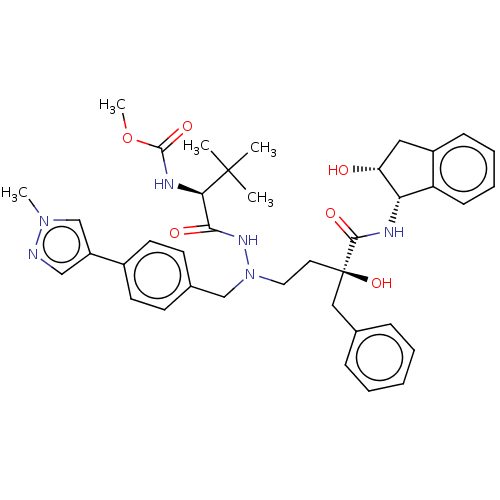
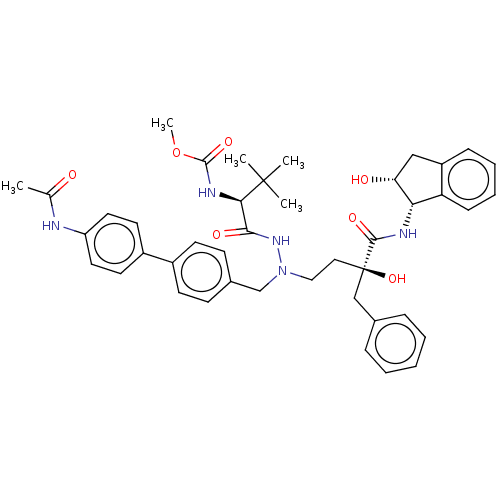
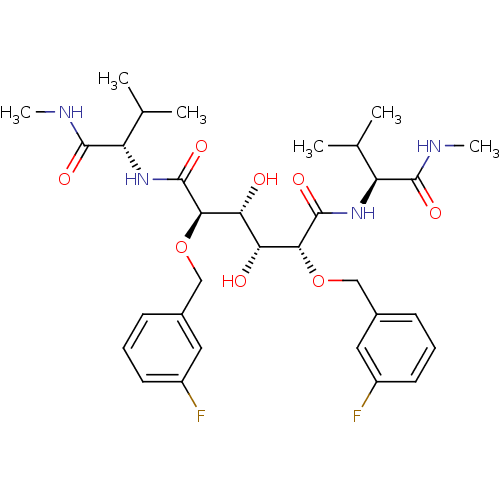
Affinity DataKi: 0.0900nM ΔG°: -58.3kJ/molepH: 5.0 T: 2°CAssay Description:Ki values were determined by using a fluorescent substrate (DABCYL-gamma-Abu-Ser-Gln-Asn-Tyr-Pro-Ile-Val-Gln-EDANS). All incubations were performed a...More data for this Ligand-Target Pair
Affinity DataKi: 0.100nM ΔG°: -58.0kJ/molepH: 5.0 T: 2°CAssay Description:A fluorimetric assay was used to determine the effects of the inhibitors on HIV-1 protease. This assay used an internally quenched fluorescent peptid...More data for this Ligand-Target Pair
Affinity DataKi: 0.100nM ΔG°: -58.0kJ/molepH: 5.0 T: 2°CAssay Description:A fluorimetric assay was used to determine the effects of the inhibitors on HIV-1 protease. This assay used an internally quenched fluorescent peptid...More data for this Ligand-Target Pair
Affinity DataKi: 0.200nMAssay Description:Inhibition of HIV1 protease expressed in Escherichia coli by fluorometric assayMore data for this Ligand-Target Pair
Affinity DataKi: 0.200nM ΔG°: -56.3kJ/molepH: 5.0 T: 2°CAssay Description:A fluorimetric assay was used to determine the effects of the inhibitors on HIV-1 protease. This assay used an internally quenched fluorescent peptid...More data for this Ligand-Target Pair
TargetGag-Pol polyprotein [489-587](Human immunodeficiency virus type 1)
LinköPing University
Curated by ChEMBL
LinköPing University
Curated by ChEMBL
Affinity DataKi: 0.200nMAssay Description:Inhibitory activity against purified HIV-1 protease expressed in E. coli in sensitive fluorometric assayMore data for this Ligand-Target Pair
Affinity DataKi: 0.300nM ΔG°: -55.3kJ/molepH: 5.0 T: 2°CAssay Description:Ki values were determined by using a fluorescent substrate (DABCYL-gamma-Abu-Ser-Gln-Asn-Tyr-Pro-Ile-Val-Gln-EDANS). All incubations were performed a...More data for this Ligand-Target Pair
Affinity DataKi: 0.300nM ΔG°: -55.3kJ/molepH: 5.0 T: 2°CAssay Description:Ki values were determined by using a fluorescent substrate (DABCYL-gamma-Abu-Ser-Gln-Asn-Tyr-Pro-Ile-Val-Gln-EDANS). All incubations were performed a...More data for this Ligand-Target Pair
Affinity DataKi: 0.300nM ΔG°: -55.3kJ/molepH: 5.0 T: 2°CAssay Description:Ki values were determined by using a fluorescent substrate (DABCYL-gamma-Abu-Ser-Gln-Asn-Tyr-Pro-Ile-Val-Gln-EDANS). All incubations were performed a...More data for this Ligand-Target Pair
Affinity DataKi: 0.300nMAssay Description:Inhibition of HIV1 protease expressed in Escherichia coli by fluorometric assayMore data for this Ligand-Target Pair
Affinity DataKi: 0.400nM ΔG°: -54.5kJ/molepH: 5.0 T: 2°CAssay Description:Ki values were determined by using a fluorescent substrate (DABCYL-gamma-Abu-Ser-Gln-Asn-Tyr-Pro-Ile-Val-Gln-EDANS). All incubations were performed a...More data for this Ligand-Target Pair
Affinity DataKi: 0.400nM ΔG°: -54.5kJ/molepH: 5.0 T: 2°CAssay Description:A fluorimetric assay was used to determine the effects of the inhibitors on HIV-1 protease. This assay used an internally quenched fluorescent peptid...More data for this Ligand-Target Pair
TargetGag-Pol polyprotein [489-587](Human immunodeficiency virus type 1)
LinköPing University
Curated by ChEMBL
LinköPing University
Curated by ChEMBL
Affinity DataKi: 0.400nMAssay Description:Inhibitory activity against purified HIV-1 protease expressed in E. coli in sensitive fluorometric assayMore data for this Ligand-Target Pair
Affinity DataKi: 0.400nM ΔG°: -54.5kJ/molepH: 5.0 T: 2°CAssay Description:A fluorimetric assay was used to determine the effects of the inhibitors on HIV-1 protease. This assay used an internally quenched fluorescent peptid...More data for this Ligand-Target Pair
Affinity DataKi: 0.5nMAssay Description:Inhibition of HIV1 protease expressed in Escherichia coli by fluorometric assayMore data for this Ligand-Target Pair
Affinity DataKi: 0.520nMAssay Description:Inhibition of HIV1 protease expressed in Escherichia coliMore data for this Ligand-Target Pair
Affinity DataKi: 0.600nM ΔG°: -53.5kJ/molepH: 5.0 T: 2°CAssay Description:Ki values were determined by using a fluorescent substrate (DABCYL-gamma-Abu-Ser-Gln-Asn-Tyr-Pro-Ile-Val-Gln-EDANS). All incubations were performed a...More data for this Ligand-Target Pair
Affinity DataKi: 0.600nM ΔG°: -53.5kJ/molepH: 5.0 T: 2°CAssay Description:Ki values were determined by using a fluorescent substrate (DABCYL-gamma-Abu-Ser-Gln-Asn-Tyr-Pro-Ile-Val-Gln-EDANS). All incubations were performed a...More data for this Ligand-Target Pair
Affinity DataKi: 0.600nM ΔG°: -53.5kJ/molepH: 5.0 T: 2°CAssay Description:A fluorimetric assay was used to determine the effects of the inhibitors on HIV-1 protease. This assay used an internally quenched fluorescent peptid...More data for this Ligand-Target Pair
Affinity DataKi: 0.700nM ΔG°: -53.1kJ/molepH: 5.0 T: 2°CAssay Description:Ki values were determined by using a fluorescent substrate (DABCYL-gamma-Abu-Ser-Gln-Asn-Tyr-Pro-Ile-Val-Gln-EDANS). All incubations were performed a...More data for this Ligand-Target Pair
Affinity DataKi: 0.800nM ΔG°: -52.8kJ/molepH: 5.0 T: 2°CAssay Description:A fluorimetric assay was used to determine the effects of the inhibitors on HIV-1 protease. This assay used an internally quenched fluorescent peptid...More data for this Ligand-Target Pair
Affinity DataKi: 0.800nMAssay Description:Inhibition of HIV1 protease expressed in Escherichia coli by fluorometric assayMore data for this Ligand-Target Pair
Affinity DataKi: 0.800nM ΔG°: -52.8kJ/molepH: 5.0 T: 2°CAssay Description:A fluorimetric assay was used to determine the effects of the inhibitors on HIV-1 protease. This assay used an internally quenched fluorescent peptid...More data for this Ligand-Target Pair
Affinity DataKi: 0.900nM ΔG°: -52.5kJ/molepH: 5.0 T: 2°CAssay Description:A fluorimetric assay was used to determine the effects of the inhibitors on HIV-1 protease. This assay used an internally quenched fluorescent peptid...More data for this Ligand-Target Pair
TargetGag-Pol polyprotein [489-587](Human immunodeficiency virus type 1)
LinköPing University
Curated by ChEMBL
LinköPing University
Curated by ChEMBL
Affinity DataKi: 0.900nMAssay Description:Inhibitory activity against purified HIV-1 protease expressed in E. coli in sensitive fluorometric assayMore data for this Ligand-Target Pair
Affinity DataKi: 0.920nM ΔG°: -52.4kJ/molepH: 5.0 T: 2°CAssay Description:A fluorimetric assay was used to determine the effects of the inhibitors on HIV-1 protease. This assay used an internally quenched fluorescent peptid...More data for this Ligand-Target Pair
Affinity DataKi: 1nMAssay Description:Inhibition of HIV1 protease expressed in Escherichia coli by fluorometric assayMore data for this Ligand-Target Pair
Affinity DataKi: 1.10nM ΔG°: -52.0kJ/molepH: 5.0 T: 2°CAssay Description:A fluorimetric assay was used to determine the effects of the inhibitors on HIV-1 protease. This assay used an internally quenched fluorescent peptid...More data for this Ligand-Target Pair
Affinity DataKi: 1.10nMAssay Description:Inhibition of HIV1 protease expressed in Escherichia coli by fluorometric assayMore data for this Ligand-Target Pair
Affinity DataKi: 1.10nMAssay Description:Inhibition of HIV1 protease expressed in Escherichia coli by fluorometric assayMore data for this Ligand-Target Pair
Affinity DataKi: 1.12nM ΔG°: -51.9kJ/molepH: 5.0 T: 2°CAssay Description:A fluorimetric assay was used to determine the effects of the inhibitors on HIV-1 protease. This assay used an internally quenched fluorescent peptid...More data for this Ligand-Target Pair
Affinity DataKi: 1.20nM ΔG°: -51.8kJ/molepH: 5.0 T: 2°CAssay Description:Ki values were determined by using a fluorescent substrate (DABCYL-gamma-Abu-Ser-Gln-Asn-Tyr-Pro-Ile-Val-Gln-EDANS). All incubations were performed a...More data for this Ligand-Target Pair
Affinity DataKi: 1.20nM ΔG°: -51.8kJ/molepH: 5.0 T: 2°CAssay Description:Ki values were determined by using a fluorescent substrate (DABCYL-gamma-Abu-Ser-Gln-Asn-Tyr-Pro-Ile-Val-Gln-EDANS). All incubations were performed a...More data for this Ligand-Target Pair
Affinity DataKi: 1.20nM ΔG°: -51.8kJ/molepH: 5.0 T: 2°CAssay Description:A fluorimetric assay was used to determine the effects of the inhibitors on HIV-1 protease. This assay used an internally quenched fluorescent peptid...More data for this Ligand-Target Pair
Affinity DataKi: 1.20nMAssay Description:Inhibition of HIV1 protease expressed in Escherichia coli by fluorometric assayMore data for this Ligand-Target Pair
Affinity DataKi: 1.22nM ΔG°: -51.7kJ/molepH: 5.0 T: 2°CAssay Description:A fluorimetric assay was used to determine the effects of the inhibitors on HIV-1 protease. This assay used an internally quenched fluorescent peptid...More data for this Ligand-Target Pair
Affinity DataKi: 1.28nM ΔG°: -51.6kJ/molepH: 5.0 T: 2°CAssay Description:A fluorimetric assay was used to determine the effects of the inhibitors on HIV-1 protease. This assay used an internally quenched fluorescent peptid...More data for this Ligand-Target Pair
Affinity DataKi: 1.40nM ΔG°: -51.4kJ/molepH: 5.0 T: 2°CAssay Description:Ki values were determined by using a fluorescent substrate (DABCYL-gamma-Abu-Ser-Gln-Asn-Tyr-Pro-Ile-Val-Gln-EDANS). All incubations were performed a...More data for this Ligand-Target Pair
Affinity DataKi: 1.40nM ΔG°: -51.4kJ/molepH: 5.0 T: 2°CAssay Description:A fluorimetric assay was used to determine the effects of the inhibitors on HIV-1 protease. This assay used an internally quenched fluorescent peptid...More data for this Ligand-Target Pair
Affinity DataKi: 1.40nMAssay Description:Inhibition of HIV1 protease expressed in Escherichia coli by fluorometric assayMore data for this Ligand-Target Pair
Affinity DataKi: 1.64nM ΔG°: -51.0kJ/molepH: 5.0 T: 2°CAssay Description:A fluorimetric assay was used to determine the effects of the inhibitors on HIV-1 protease. This assay used an internally quenched fluorescent peptid...More data for this Ligand-Target Pair
Affinity DataKi: 1.65nM ΔG°: -51.0kJ/molepH: 5.0 T: 2°CAssay Description:A fluorimetric assay was used to determine the effects of the inhibitors on HIV-1 protease. This assay used an internally quenched fluorescent peptid...More data for this Ligand-Target Pair
Affinity DataKi: 1.70nMAssay Description:Inhibition of HIV1 protease expressed in Escherichia coli by fluorometric assayMore data for this Ligand-Target Pair
Affinity DataKi: 1.70nMAssay Description:Inhibition of HIV1 protease expressed in Escherichia coli by fluorometric assayMore data for this Ligand-Target Pair
Affinity DataKi: 1.70nMAssay Description:Inhibition of HIV1 protease expressed in Escherichia coli by fluorometric assayMore data for this Ligand-Target Pair
Affinity DataKi: 1.73nM ΔG°: -50.8kJ/molepH: 5.0 T: 2°CAssay Description:A fluorimetric assay was used to determine the effects of the inhibitors on HIV-1 protease. This assay used an internally quenched fluorescent peptid...More data for this Ligand-Target Pair
Affinity DataKi: 1.79nM ΔG°: -50.8kJ/molepH: 5.0 T: 2°CAssay Description:A fluorimetric assay was used to determine the effects of the inhibitors on HIV-1 protease. This assay used an internally quenched fluorescent peptid...More data for this Ligand-Target Pair
Affinity DataKi: 1.80nMAssay Description:Inhibition of HIV1 protease expressed in Escherichia coli by fluorometric assayMore data for this Ligand-Target Pair
Affinity DataKi: 1.83nM ΔG°: -50.7kJ/molepH: 5.0 T: 2°CAssay Description:A fluorimetric assay was used to determine the effects of the inhibitors on HIV-1 protease. This assay used an internally quenched fluorescent peptid...More data for this Ligand-Target Pair
Affinity DataKi: 1.92nM ΔG°: -50.6kJ/molepH: 5.0 T: 2°CAssay Description:A fluorimetric assay was used to determine the effects of the inhibitors on HIV-1 protease. This assay used an internally quenched fluorescent peptid...More data for this Ligand-Target Pair

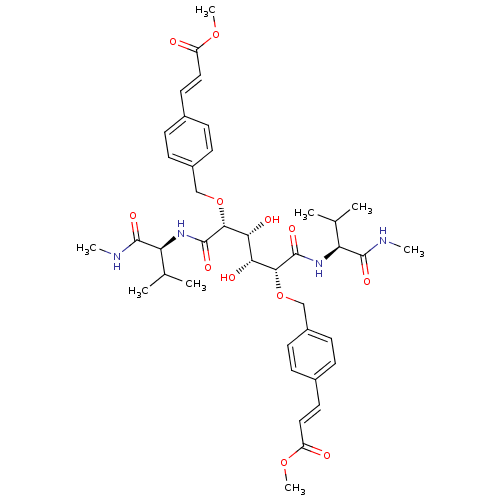
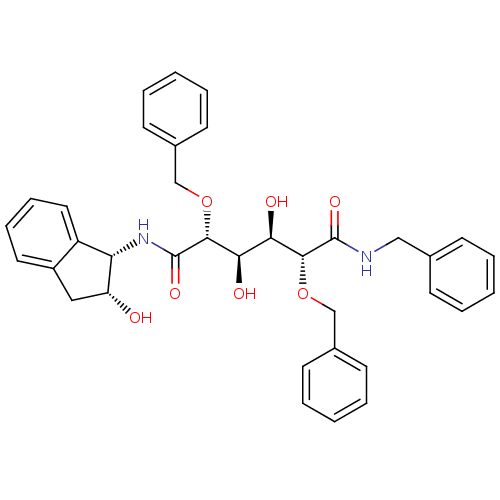
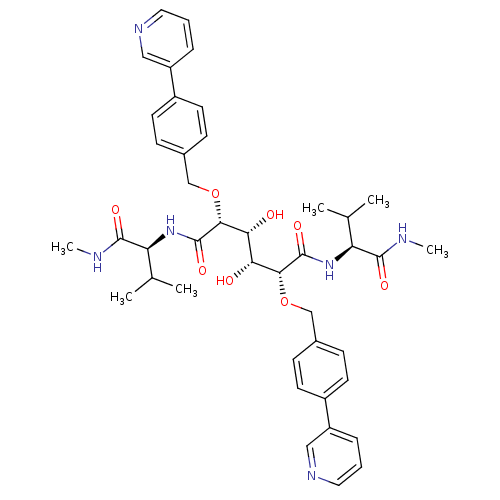
 3D Structure (crystal)
3D Structure (crystal)