Activity Spreadsheet -- Enzyme Inhibition Constant Data from BindingDB (change energy unit to kcal/mol)
 Found 10 hits in this display
Found 10 hits in this display
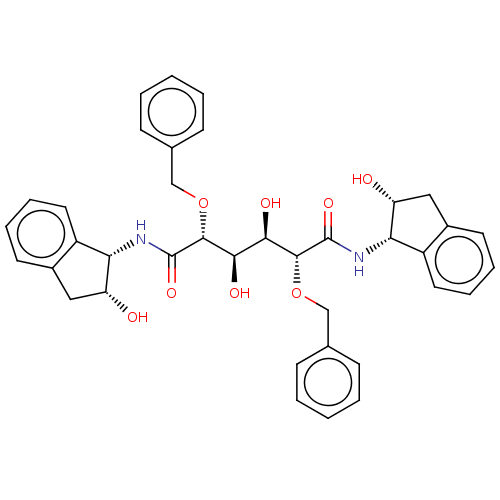 BDBM358((2R,3R,4R,5R)-2,5-bis(benzyloxy)-3,4-dihydroxy-N,N...)
BDBM358((2R,3R,4R,5R)-2,5-bis(benzyloxy)-3,4-dihydroxy-N,N...)
 BDBM358((2R,3R,4R,5R)-2,5-bis(benzyloxy)-3,4-dihydroxy-N,N...)
BDBM358((2R,3R,4R,5R)-2,5-bis(benzyloxy)-3,4-dihydroxy-N,N...)
Protease(Human immunodeficiency virus 1 (HIV-1))Stockholm University
Curated by ChEMBL
 BDBM358((2R,3R,4R,5R)-2,5-bis(benzyloxy)-3,4-dihydroxy-N,N...)
BDBM358((2R,3R,4R,5R)-2,5-bis(benzyloxy)-3,4-dihydroxy-N,N...)
 BDBM358((2R,3R,4R,5R)-2,5-bis(benzyloxy)-3,4-dihydroxy-N,N...)
BDBM358((2R,3R,4R,5R)-2,5-bis(benzyloxy)-3,4-dihydroxy-N,N...)
 BDBM358((2R,3R,4R,5R)-2,5-bis(benzyloxy)-3,4-dihydroxy-N,N...)
BDBM358((2R,3R,4R,5R)-2,5-bis(benzyloxy)-3,4-dihydroxy-N,N...)
 BDBM358((2R,3R,4R,5R)-2,5-bis(benzyloxy)-3,4-dihydroxy-N,N...)
BDBM358((2R,3R,4R,5R)-2,5-bis(benzyloxy)-3,4-dihydroxy-N,N...)
Protease(Human immunodeficiency virus 1 (HIV-1))Stockholm University
Curated by ChEMBL
Protease(Human immunodeficiency virus 1 (HIV-1))Stockholm University
Curated by ChEMBL
Protease(Human immunodeficiency virus 1 (HIV-1))Stockholm University
Curated by ChEMBL