Query String: BPV pic
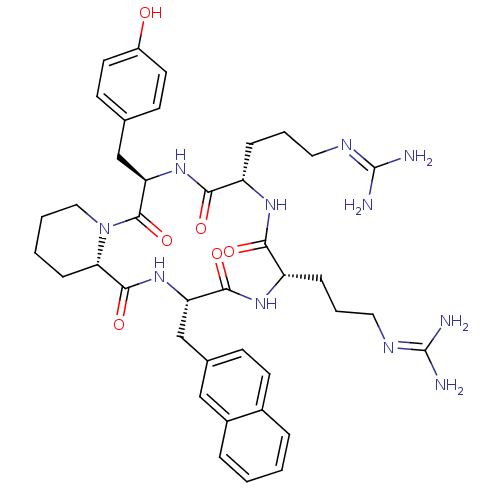 CHEMBL218863 cyclo(-D-Tyr-L-Arg-L-Arg-L-Nal-L-Pic-) BDBM50202370
CHEMBL218863 cyclo(-D-Tyr-L-Arg-L-Arg-L-Nal-L-Pic-) BDBM50202370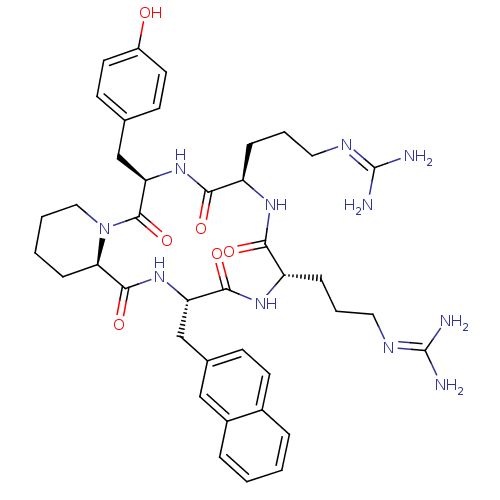 CHEMBL218864 cyclo(-D-Tyr-D-Arg-L-Arg-L-Nal-D-Pic-) BDBM50202342
CHEMBL218864 cyclo(-D-Tyr-D-Arg-L-Arg-L-Nal-D-Pic-) BDBM50202342 CHEMBL375168 cyclo(-D-Tyr-L-Arg-L-Arg-L-Nal-D-Pic-) BDBM50202372
CHEMBL375168 cyclo(-D-Tyr-L-Arg-L-Arg-L-Nal-D-Pic-) BDBM50202372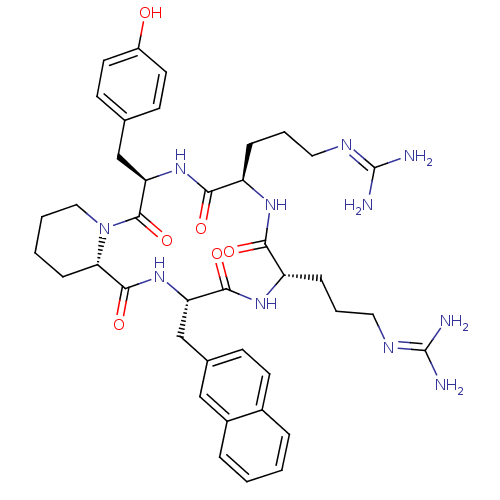 cyclo(-D-Tyr-D-Arg-L-Arg-L-Nal-L-Pic-) CHEMBL373637 BDBM50202353
cyclo(-D-Tyr-D-Arg-L-Arg-L-Nal-L-Pic-) CHEMBL373637 BDBM50202353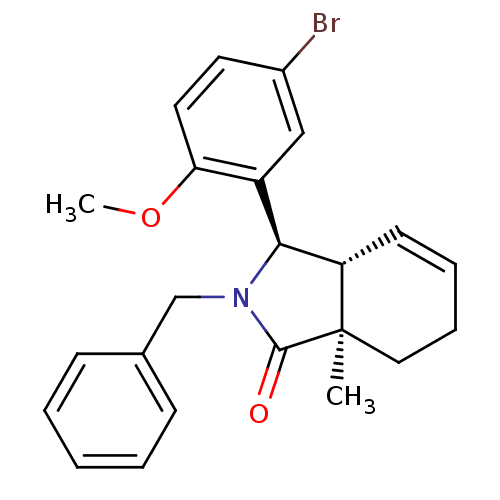 (3R,3aR,7aR)-2-benzyl-3-(5-bromo-2-methoxy-phenyl)-7a-methyl-3,3a,6,7-tetrahydroisoindol-1-one (3R,3aR,7aR)-3-(5-bromanyl-2-methoxy-phenyl)-7a-methyl-2-(phenylmethyl)-3,3a,6,7-tetrahydroisoindol-1-one SMR000465410 MLS000882796 (3R,3aR,7aR)-3-(5-bromo-2-methoxyphenyl)-7a-methyl-2-(phenylmethyl)-3,3a,6,7-tetrahydroisoindol-1-one BDBM53403 PiC cid_16060470 (3R,3aR,7aR)-2-benzyl-3-(5-bromo-2-methoxyphenyl)-7a-methyl-3,3a,6,7-tetrahydroisoindol-1-one
(3R,3aR,7aR)-2-benzyl-3-(5-bromo-2-methoxy-phenyl)-7a-methyl-3,3a,6,7-tetrahydroisoindol-1-one (3R,3aR,7aR)-3-(5-bromanyl-2-methoxy-phenyl)-7a-methyl-2-(phenylmethyl)-3,3a,6,7-tetrahydroisoindol-1-one SMR000465410 MLS000882796 (3R,3aR,7aR)-3-(5-bromo-2-methoxyphenyl)-7a-methyl-2-(phenylmethyl)-3,3a,6,7-tetrahydroisoindol-1-one BDBM53403 PiC cid_16060470 (3R,3aR,7aR)-2-benzyl-3-(5-bromo-2-methoxyphenyl)-7a-methyl-3,3a,6,7-tetrahydroisoindol-1-one
- ChEMBL_490891 (CHEMBL989167) Displacement of [125I]PIC from human imidazoline receptor 1 expressed in rat PC12 cells
- ChEMBL_1583798 (CHEMBL3815885) Displacement of [125I]PIC from Imidazoline-1 receptor in rat PC12 cell membrane by gamma counting method
- ChEMBL_1467221 (CHEMBL3411418) Displacement of [125I] PIC from I1 imidazoline receptor in rat PC12 cells after 45 mins by gamma counting
- ChEMBL_490890 (CHEMBL989166) Displacement of [125I]PIC from human imidazoline receptor 1 in human platelets analyzed under norepinephrine mask of alpha 2AR
- ChEMBL_1583797 (CHEMBL3815884) Displacement of [125I]PIC from Imidazoline-1 receptor in rat PC12 cell membrane incubated for 30 mins by gamma counting method
- ChEMBL_1911577 (CHEMBL4414023) Inhibition of human 6His-tagged SHP2 (1 to 525 residues) expressed in Escherichia coli BL21 Star (DE3) using DiFMUP as surrogate substrate as preincubated for 30 to 60 mins followed by substrate addition and measured after 30 mins bpV-based fluorescence assay
- In vitro degradation assay In vitro degradation assay protocol for compounds 3-33: Panc02.13 cells were purchased from ATCC and cultured in RPMI-1640 (Gibco), supplemented with 15% FBS (ATCC) and 10 Units/mL human recombinant insulin (Gibco). PROTAC treatments were carried out in 12-well plates for 16 h. TLR3 agonist Poly I:C (Invivogen; tlr1-pic) was added for the final 3 h. Cells were harvested, and lysed in RIPA buffer (50 mM Tris pH8, 150 mM NaCl, 1% Tx-100, 0.1% SDS, 0.5% Sodium Deoxycholate) supplemented with protease and phosphatase inhibitors. Lysates were clarified at 16,000 g for 10 minutes, and supernatants were separated by SDS-PAGE. Immunoblotting was performed using standard protocols. The antibodies used were TBK1 (Cell Signaling#3504), pIRF3 (abcam#ab76493), and GAPDH (Cell Signaling#5174).
- Inhibition Assay The inhibition of SHP2 by compounds of the invention (concentrations varying from 0.003-100 μM) was monitored using an assay in which 0.5 nM of SHP2 was incubated with of 0.5 μM of peptide IRS 1_pY1172(dPEG8)pY1222 (sequence: H2N-LN(pY)IDLDLV(dPEG8)LST(pY)ASINFQK-amide) (SEQ ID NO:1 and SEQ ID NO: 2, respectively, in order of appearance). After 30-60 minutes incubation at 25° C., the surrogate substrate DiFMUP (Invitrogen, cat #D6567) was added to the reaction and incubated at 25° C. for 30 minutes. The reaction was then quenched by the addition of 5 μl of a 160 μM solution of bpV(Phen) (Enzo Life Sciences cat #ALX-270-204). The fluorescence signal was monitored using a microplate reader (Envision, Perki-Elmer) using excitation and emission wavelengths of 340 nm and 450 nm, respectively. The inhibitor dose response curves were analyzed using normalized IC50 regression curve fitting with control based normalization.
- SHP2 Phosphatase Assay IC50 values were determined at room temperature in 384-well black polystyrene plate, using a final reaction volume of 15 μl and the following assay buffer conditions: 60 mM Hepes (pH=7.2), 75 mM NaCl, 75 mM KCl, and 1 mM EDTA, 0.05% P-20, 5 mM dithiothreitol (DTT). Full length SHP2 enzyme (diluted to 0.1 nM in reaction buffer) were co-incubated with 1 uM IRS-1 peptide and 0.01 nM to 10 μM compounds of the disclosure for 60 min. The surrogate substrate DiFMUP (5 μL, 100 LIM) was added, and incubated at rt for 60 min. The reaction was then quenched by the addition of 5 μL of a 40 μM solution of bpV(Phen). The fluorescence signal was monitored using a microplate reader (Envision, Perkin-Elmer) using excitation and emission wavelengths of 360 nm and 450 nm, respectively. The inhibitor dose-response curves were analyzed using normalized IC50 regression curve fitting with control-based normalization.
- SHP2 Allosteric Inhibition Assay More specifically, the phosphatase reactions were performed at room temperature in 384-well black polystyrene plate, flat bottom, low flange, non-binding surface (Corning, Cat #3575) using a final reaction volume of 25 μL and the following assay buffer conditions: 60 mM HEPES, pH 7.2, 75 mM NaCl, 75 mM KCl, 1 mM EDTA, 0.05% P-20, 5 mM DTT.The inhibition of SHP2 (concentrations varying from 0.003-100 μM) was monitored using an assay in which 0.5 nM of SHP2 was incubated with of 0.5 μM of peptide IRS1_pY1172(dPEG8)pY1222 (sequence: H2N-LN(pY)IDLDLV(dPEG8)LST(pY)ASINFQK-amide (SEQ ID NO: 1)). After 30-60 minutes incubation at 25° C., the surrogate substrate DiFMUP (Invitrogen, cat #D6567) was added to the reaction and incubated at 25° C. for 30 minutes. The reaction was then quenched by the addition of 5 μL of a 160 μM solution of bpV(Phen) (Enzo Life Sciences cat #ALX-270-204). The fluorescence signal was monitored using a microplate reader (Envision, Perki-Elmer) using excitation and emission wavelengths of 340 nm and 450 nm, respectively. The inhibitor dose response curves were analyzed using normalized IC50 regression curve fitting with control based normalization.
- SHP2 Allosteric Inhibition Assay More specifically, the phosphatase reactions were performed at room temperature in 384-well black polystyrene plate, flat bottom, low flange, non-binding surface (Corning, Cat #3575) using a final reaction volume of 25 μL and the following assay buffer conditions: 60 mM HEPES, pH 7.2, 75 mM NaCl, 75 mM KCl, 1 mM EDTA, 0.05% P-20, 5 mM DTT.The inhibition of SHP2 by compounds of the invention (concentrations varying from 0.003-100 μM) was monitored using an assay in which 0.5 nM of SHP2 was incubated with of 0.5 μM of peptide IRS1_pY1172(dPEG8)pY1222 (sequence: H2N-LN(pY)IDLDLV(dPEG8)LST(pY)ASINFQK-amide) (SEQ ID NO:1). After 30-60 minutes incubation at 25° C., the surrogate substrate DiFMUP (Invitrogen, cat #D6567) was added to the reaction and incubated at 25° C. for 30 minutes. The reaction was then quenched by the addition of 5 μl of a 160 μM solution of bpV(Phen) (Enzo Life Sciences cat #ALX-270-204). The fluorescence signal was monitored using a microplate reader (Envision, Perki-Elmer) using excitation and emission wavelengths of 340 nm and 450 nm, respectively. The inhibitor dose response curves were analyzed using normalized IC50 regression curve fitting with control based normalization.
- SHP2 Allosteric Inhibition Assay More specifically, the phosphatase reactions were performed at room temperature in 384-well black polystyrene plate, flat bottom, low flange, non-binding surface (Corning, Cat#3575) using a final reaction volume of 25 μL and the following assay buffer conditions: 60 mM HEPES, pH 7.2, 75 mM NaCl, 75 mM KCl, 1 mM EDTA, 0.05% P-20, 5 mM DTT. The inhibition of SHP2 by compounds of the invention (concentrations varying from 0.003-100 μM) was monitored using an assay in which 0.5 nM of SHP2 was incubated with of 0.5 μM of peptide IRS1_pY1172(dPEG8)pY1222 (sequence: H2N-LN(pY)IDLDLV(dPEG8)LST(pY)ASINFQK-amide) (SEQ ID NO:1). After 30-60 minutes incubation at 25° C., the surrogate substrate DiFMUP (Invitrogen, cat# D6567) was added to the reaction and incubated at 25° C. for 30 minutes. The reaction was then quenched by the addition of 5 μl of a 160 μM solution of bpV(Phen) (Enzo Life Sciences cat# ALX-270-204). The fluorescence signal was monitored using a microplate reader (Envision, Perki-Elmer) using excitation and emission wavelengths of 340 nm and 450 nm, respectively. The inhibitor dose response curves were analyzed using normalized IC50 regression curve fitting with control based normalization.
- SHP2 Allosteric Inhibition Assay The phosphatase reaction was conducted in 384-well black polystyrene plates (Corning, Cat #3575) with flat bottom, low edge and non-binding surface at room temperature and 25 μl final volume of the following buffer condition: 60 mM HEPES, pH 7.2, 75 mM NaCl, 75 mM KCl, 1 mM EDTA, 0.05% P-20, 5 mM DTT.The following experiments were conducted to monitor the SHP2 inhibition by the compounds (at concentrations of 0.0003-100 μM) in this invention:Wherein, incubate 0.5 nM SHP2 with 0.5 μM peptide IRS1_pY1172 (dPEG8) pY1222 (sequence: H2N-LN (pY) IDLDLV (dPEG8) LST (pY) ASINFQK-amide) (SEQ ID NO: 1) (WO2016/203406A1). After incubation at 25° C. for 30-60 minutes, the alternative substrate DiFMUP (Invitrogen, cat #D6567) was added to the reaction and incubated at 25° C. for 30 minutes. The reaction was then carefully diluted by adding 5 μL of a 160 μM bpV (Phen) solution (Enzo Life Sciences cat #ALX-270-204). The fluorescence signal was monitored using a microplate reader (VARIOSKAN LUX, Thermo) with excitation and emission wavelengths of 340 nm and 450 nm, respectively. The inhibitory dose-response curve is analyzed using a standardized IC50 regression curve based on control-based normalization.
- SHP2 Inhibition Enzymatic Assay Assay volume of 20 μL/well was assembled in 384 well black polystyrene low-binding microplates (Greiner), using the following buffer: 60 mM HEPES pH 7.2, 75 mM NaCl, 75 mM KCl, 1 mM EDTA pH 8, 0.05% tween-20, 5 mM DTT. The SHP-2 enzyme (synthetized by Origene, Met1-Leu525, cat #TP750155) was used at a final concentration of 0.5 nM. The enzyme was activated by 500 nM IRS1 peptide (sequence: H2N-LN(pY)IDLDLV(dPEG8)LST(pY)ASINFQK-amide SEQ ID No. 1) and incubated with 75 μM DiFMUP (Sigma) as substrate.Briefly, DMSO serially diluted testing compounds were transferred to the bottom of the assay plate. SHP2 was then added together with the IRS1 peptide. 30 min post incubation, the DiFMUP substrate was added to the reaction and incubated 30 min at room temperature. Finally 5 μL of 160 μM bpV (Potassium bisperoxo[1,10-phenanthroline]oxovanadate [V], Sigma) were added to stop and quench the reaction. The fluorescence was detected by a microplate reader (Envision, PerkinElmer) according to the DiFMUP excitation and emission wavelength. The lower the fluorescence the higher the SHP2 inhibition. The activity of each compound dilution was calculated as percentage of inhibition between vehicle (DMSO, 0% inhibition) and no enzyme (100% inhibition). The percentage inhibition is fitted against the compound dilutions with a four-parameter logistic regression.
- Fluorescence Assay for Recombinant Human (RH) DPP1 The activity of DPP1 was determined by measuring the enzymatic release of amino methyl coumarin (AMC) from the peptide substrate (H-Gly-Arg-AMC), which leads to an increase in fluorescence intensity at λex=350 nm and λem=450 nm. The assay was carried out in black 384 well plates in a final volume of 10 μl at rt. The assay conditions contained the following: 25 mM piperazine buffer pH 5.0; 50 mM NaCl, 5 mM DTT; 0.005 (v/v) Triton X-100; 50 μM H-Gly-Arg-AMC and 96.4 pM rhDPP1 Potential inhibitors were diluted in DMSO to generate 100× of the final assay concentration. The compounds were tested at 10 concentrations with half-log dilution steps (highest concentration typically 1 μM) and with a final DMSO concentration of 1% (v/v). Routinely, inhibitors were pre-incubated with rhDPP1 for 30 min prior to the addition of the peptide substrate to start the reaction for a further 30 min. After incubation the plates were read in a fluorescence plate reader using the above emission and excitation wavelengths. The pIC50 were determined using a 4-paramater logistic equation in a non-linear curve fitting routine (Smartfit, Genedata Screener ). A standard DPP1 inhibitor, 4-amino-N-[(1S)-1-cyano-2-(4′-cyanobiphenyl-4-yl)ethyl]tetrahydro-2H-pyran-4-carboxamide (WO2010/128324, Ex. 3) was used as a positive control and 1% (v/v) DMSO was used as a negative control in the assay.
- Allosteric Inhibition Assay SHP2 is allosterically activated through binding of bis-tyrosyl-phosphorylated peptides to its Src Homology 2 (SH2) domains. The latter activation step leads to the release of the auto-inhibitory interface of SHP2, which in turn renders the SHP2 protein tyrosine phosphatase (PTP) active and available for substrate recognition and reaction catalysis. The catalytic activity of SHP2 was monitored using the surrogate substrate DiFMUP in a prompt fluorescence assay format.More specifically, the phosphatase reactions were performed at room temperature in 384-well black polystyrene plate, flat bottom, low flange, non-binding surface (Corning, Cat #3575) using a final reaction volume of 25 μL and the following assay buffer conditions: 60 mM HEPES, pH 7.2, 75 mM NaCl, 75 mM KCl, 1 mM EDTA, 0.05% P-20, 5 mM DTT.The inhibition of SHP2 by compounds of the invention (concentrations varying from 0.003-100 μM) was monitored using an assay in which 0.5 nM of SHP2 was incubated with of 0.5 μM of peptide IRS1_pY1172(dPEG8)pY1222 (sequence: H2N-LN(pY)IDLDLV(dPEG8)LST(pY)ASINFQK-amide) (SEQ ID NO:1). After 30-60 minutes incubation at 25° C., the surrogate substrate DiFMUP (Invitrogen, cat #D6567) was added to the reaction and incubated at 25° C. for 30 minutes. The reaction was then quenched by the addition of 5 μl of a 160 μM solution of bpV(Phen) (Enzo Life Sciences cat #ALX-270-204). The fluorescence signal was monitored using a microplate reader (Envision, Perki-Elmer) using excitation and emission wavelengths of 340 nm and 450 nm, respectively. The inhibitor dose response curves were analyzed using normalized IC50 regression curve fitting with control based normalization.
- Back Scattering Interferometry Technology (BSI) To measure the direct binding of small molecules with CFTR, the interaction between small molecules and membrane fragments derived from HEK293 cells over-expressing WT CFTR was investigated on the TruBind BSI System.HEK293 containing WT CFTR and HEK293 control membrane fractions were prepared as follows. HEK293 were transiently transfected with WT CFTR or left untreated, washed with PBS and collected in cold PBS supplemented with protease inhibitor cocktail (PIC). Cells were centrifuged and resuspended in homogenisation buffer (15 mM Tris-HCl pH 7.5, 2 mM MgCl2, 0.3 mM EDTA, 1 mM EGTA+protease inhibitors). For compound testing HEK293 WT CFTR or HEK293 membrane fractions, at final concentration of 10 μg/mL in 50 mM Tris-HCl pH 7.5, 1 mM EDTA with 1.2% DMSO were mixed 1/1 with a serial dilution of the compound, starting from 10 μM, in 50 mM Tris-HCl pH 7.5, 1 mM EDTA with 1.2% DMSO. Mixtures were incubated at room temperature for 4 hours before being run on the BSI instrument. Samples were measured in quadruplicate in dual channel mode which allows the simultaneous measurement of specific, WT CFTR membranes (assay), as well as unspecific, control membranes (reference). For each assay the reference data is subtracted, point by point, from the assay data and plotted as fringe shift in units of radions. Each compound was run to have at least two successful experiments with good reproducibility. Success was defined as having a binding signal with a R2>0.7. The final data is exported to Graphpad Prism and fit with a one-site binding equation to determine a Kd for the assay. The data obtained using the method demonstrates clearly that the test compound as defined in the present claims directly bind to CFTR with low Kd.
- SHP2 Allosteric Inhibition Assay SHP2 is allosterically activated through binding of bis-tyrosyl-phorphorylated peptides to its Src Homology 2 (SH2) domains. The latter activation step leads to the release of the auto-inhibitory interface of SHP2, which in turn renders the SHP2 protein tyrosine phosphatase (PTP) active and available for substrate recognition and reaction catalysis. The catalytic activity of SHP2 was monitored using the surrogate substrate DiFMUP in a prompt fluorescence assay format.More specifically, the phosphatase reactions were performed at room temperature in 384-well black polystyrene plate, flat bottom, low flange, non-binding surface (Corning, Cat#3575) using a final reaction volume of 25 μL and the following assay buffer conditions: 60 mM HEPES, pH 7.2, 75 mM NaCl, 75 mM KCl, 1 mM EDTA, 0.05% P-20, 5 mM DTT.The inhibition of SHP2 by compounds of the invention (concentrations varying from 0.003-100 μM) was monitored using an assay in which 0.5 nM of SHP2 was incubated with of 0.5 μM of peptide IRS1_pY1172(dPEG8)pY1222 (sequence: H2N-LN(pY)IDLDLV(dPEG8)LST(pY)ASINFQK-amide) (SEQ ID NO: 1). After 30-60 minutes incubation at 25° C., the surrogate substrate DiFMUP (Invitrogen, cat# D6567) was added to the reaction and incubated at 25° C. for 30 minutes. The reaction was then quenched by the addition of 5 μl of a 160 μM solution of bpV(Phen) (Enzo Life Sciences cat# ALX-270-204). The fluorescence signal was monitored using a microplate reader (Envision, Perki-Elmer) using excitation and emission wavelengths of 340 nm and 450 nm, respectively. The inhibitor dose response curves were analyzed using normalized IC50 regression curve fitting with control based normalization.
- SHP2 Allosteric Inhibition Assay SHP2 is allosterically activated through binding of bis-tyrosyl-phorphorylated peptides to its Src Homology 2 (SH2) domains. The latter activation step leads to the release of the auto-inhibitory interface of SHP2, which in turn renders the SHP2 protein tyrosine phosphatase (PTP) active and available for substrate recognition and reaction catalysis. The catalytic activity of SHP2 was monitored using the surrogate substrate DiFMUP in a prompt fluorescence assay format.More specifically, the phosphatase reactions were performed at room temperature in 384-well black polystyrene plate, flat bottom, low flange, non-binding surface (Corning, Cat#3575) using a final reaction volume of 25 μL and the following assay buffer conditions: 60 mM HEPES, pH 7.2, 75 mM NaCl, 75 mM KCl, 1 mM EDTA, 0.05% P-20, 5 mM DTT.The inhibition of SHP2 by compounds of the invention (concentrations varying from 0.003-100 μM) was monitored using an assay in which 0.5 nM of SHP2 was incubated with of 0.5 μM of peptide IRS1_pY1172(dPEG8)pY1222 (sequence: H2N-LN(pY)IDLDLV(dPEG8)LST(pY)ASINFQK-amide) (SEQ ID NO:1). After 30-60 minutes incubation at 25° C., the surrogate substrate DiFMUP (Invitrogen, cat# D6567) was added to the reaction and incubated at 25° C. for 30 minutes. The reaction was then quenched by the addition of 5 μl of a 160 μM solution of bpV(Phen) (Enzo Life Sciences cat# ALX-270-204). The fluorescence signal was monitored using a microplate reader (Envision, Perki-Elmer) using excitation and emission wavelengths of 340 nm and 450 nm, respectively. The inhibitor dose response curves were analyzed using normalized IC50 regression curve fitting with control based normalization.
- SHP2 Allosteric Inhibition Assay SHP2 is allosterically activated through binding of bis-tyrosyl-phorphorylated peptides to its Src Homology 2 (SH2) domains. The latter activation step leads to the release of the auto-inhibitory interface of SHP2, which in turn renders the SHP2 protein tyrosine phosphatase (PTP) active and available for substrate recognition and reaction catalysis. The catalytic activity of SHP2 was monitored using the surrogate substrate DiFMUP in a prompt fluorescence assay format.More specifically, the phosphatase reactions were performed at room temperature in 384-well black polystyrene plate, flat bottom, low flange, non-binding surface (Corning, Cat#3575) using a final reaction volume of 25 μL and the following assay buffer conditions: 60 mM HEPES, pH 7.2, 75 mM NaCl, 75 mM KCl, 1 mM EDTA, 0.05% P-20, 5 mM DTT.The inhibition of SHP2 by compounds of the invention (concentrations varying from 0.003-100 μM) was monitored using an assay in which 0.5 nM of SHP2 was incubated with of 0.5 μM of peptide IRS1_pY1172(dPEG8)pY1222 (sequence: H2N-LN(pY)IDLDLV(dPEG8)LST(pY)ASINFQK-amide) (SEQ ID NO:1). After 30-60 minutes incubation at 25° C., the surrogate substrate DiFMUP (Invitrogen, cat#D6567) was added to the reaction and incubated at 25° C. for 30 minutes. The reaction was then quenched by the addition of 5 μl of a 160 μM solution of bpV(Phen) (Enzo Life Sciences cat#ALX-270-204). The fluorescence signal was monitored using a microplate reader (Envision, Perki-Elmer) using excitation and emission wavelengths of 340 nm and 450 nm, respectively. The inhibitor dose response curves were analyzed using normalized IC50 regression curve fitting with control based normalization.
- SHP2 Allosteric Inhibition Assay SHP2 is allosterically activated through binding of bis-tyrosyl-phosphorylated peptides to its Src Homology 2 (SH2) domains. The latter activation step leads to the release of the auto-inhibitory interface of SHP2, which in turn renders the SHP2 protein tyrosine phosphatase (PTP) active and available for substrate recognition and reaction catalysis. The catalytic activity of SHP2 was monitored using the surrogate substrate DiFMUP in a prompt fluorescence assay format.More specifically, the phosphatase reactions were performed at room temperature in 384-well black polystyrene plate, flat bottom, low flange, non-binding surface (Corning, Cat#3575) using a final reaction volume of 25 μL and the following assay buffer conditions: 60 mM HEPES, pH 7.2, 75 mM NaCl, 75 mM KCl, 1 mM EDTA, 0.05% P-20, 5 mM DTT.The inhibition of SHP2 by compounds of the invention (concentrations varying from 0.003-100 μM) was monitored using an assay in which 0.5 nM of SHP2 was incubated with of 0.5 μM of peptide IRS1_pY1172(dPEG8)pY1222 (sequence: H2N-LN(pY)IDLDLV(dPEG8)LST(pY)ASINFQK-amide) (SEQ ID NO:1). After 30-60 minutes incubation at 25° C., the surrogate substrate DiFMUP (Invitrogen, cat# D6567) was added to the reaction and incubated at 25° C. for 30 minutes. The reaction was then carefully diluted by the addition of 5 μL of a 160 μM solution of bpV(Phen) (Enzo Life Sciences cat# ALX-270-204). The fluorescence signal was monitored using a microplate reader (Envision, Perki-Elmer) using excitation and emission wavelengths of 340 nm and 450 nm, respectively. The inhibitor dose response curves were analyzed using normalized IC50 regression curve fitting with control based normalization.
 CHEMBL218863 cyclo(-D-Tyr-L-Arg-L-Arg-L-Nal-L-Pic-) BDBM50202370
CHEMBL218863 cyclo(-D-Tyr-L-Arg-L-Arg-L-Nal-L-Pic-) BDBM50202370 CHEMBL218864 cyclo(-D-Tyr-D-Arg-L-Arg-L-Nal-D-Pic-) BDBM50202342
CHEMBL218864 cyclo(-D-Tyr-D-Arg-L-Arg-L-Nal-D-Pic-) BDBM50202342 CHEMBL375168 cyclo(-D-Tyr-L-Arg-L-Arg-L-Nal-D-Pic-) BDBM50202372
CHEMBL375168 cyclo(-D-Tyr-L-Arg-L-Arg-L-Nal-D-Pic-) BDBM50202372 cyclo(-D-Tyr-D-Arg-L-Arg-L-Nal-L-Pic-) CHEMBL373637 BDBM50202353
cyclo(-D-Tyr-D-Arg-L-Arg-L-Nal-L-Pic-) CHEMBL373637 BDBM50202353 (3R,3aR,7aR)-2-benzyl-3-(5-bromo-2-methoxy-phenyl)-7a-methyl-3,3a,6,7-tetrahydroisoindol-1-one (3R,3aR,7aR)-3-(5-bromanyl-2-methoxy-phenyl)-7a-methyl-2-(phenylmethyl)-3,3a,6,7-tetrahydroisoindol-1-one SMR000465410 MLS000882796 (3R,3aR,7aR)-3-(5-bromo-2-methoxyphenyl)-7a-methyl-2-(phenylmethyl)-3,3a,6,7-tetrahydroisoindol-1-one BDBM53403 PiC cid_16060470 (3R,3aR,7aR)-2-benzyl-3-(5-bromo-2-methoxyphenyl)-7a-methyl-3,3a,6,7-tetrahydroisoindol-1-one
(3R,3aR,7aR)-2-benzyl-3-(5-bromo-2-methoxy-phenyl)-7a-methyl-3,3a,6,7-tetrahydroisoindol-1-one (3R,3aR,7aR)-3-(5-bromanyl-2-methoxy-phenyl)-7a-methyl-2-(phenylmethyl)-3,3a,6,7-tetrahydroisoindol-1-one SMR000465410 MLS000882796 (3R,3aR,7aR)-3-(5-bromo-2-methoxyphenyl)-7a-methyl-2-(phenylmethyl)-3,3a,6,7-tetrahydroisoindol-1-one BDBM53403 PiC cid_16060470 (3R,3aR,7aR)-2-benzyl-3-(5-bromo-2-methoxyphenyl)-7a-methyl-3,3a,6,7-tetrahydroisoindol-1-one