 DASATINIB BDBM31089
DASATINIB BDBM31089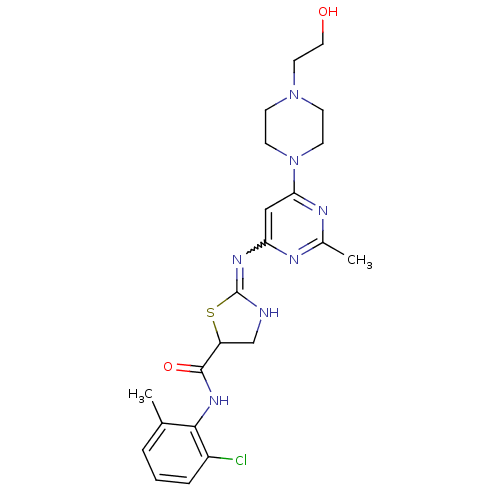 Dasatinib BDBM82130
Dasatinib BDBM82130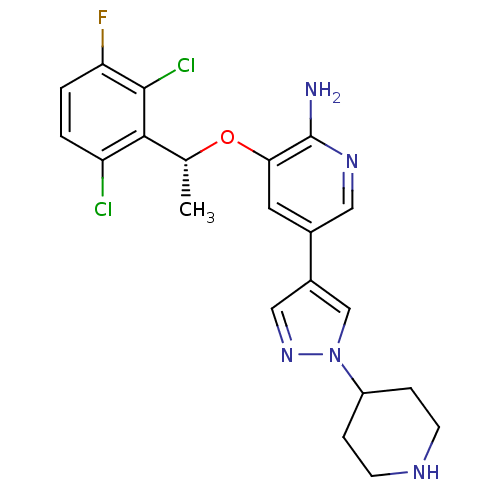 US12129258, Example Crizotinib BDBM50306682 US10370379, Crizotinib (R)-3-(1-(2,6-dichloro-3-fluorophenyl)ethoxy)-5-(1-(piperidin-4-yl)-1H-pyrazol-4-yl)pyridin-2-amine CHEMBL601719 US11059827, Compound Crizotinib US9199944, Crizotinib PF-2341066 3-[(1R)-1-(2,6-dichloro-3-fluorophenyl)ethoxy]-5-(1-piperidin-4-yl-1H-pyrazol-4-yl)pyridin-2-amine US10543199, Compound Crizotinib US9126941, PF-2341066 US11517561, Compound Crizotinib CRIZOTINIB 3-(2,6-dichloro-3-fluorobenzyloxy)-5-(1-(piperidin-4-yl)-1H-pyrazol-4-yl)pyridin-2-amine US10780082, Compound Crizotinib US9226923, Crizotinib
US12129258, Example Crizotinib BDBM50306682 US10370379, Crizotinib (R)-3-(1-(2,6-dichloro-3-fluorophenyl)ethoxy)-5-(1-(piperidin-4-yl)-1H-pyrazol-4-yl)pyridin-2-amine CHEMBL601719 US11059827, Compound Crizotinib US9199944, Crizotinib PF-2341066 3-[(1R)-1-(2,6-dichloro-3-fluorophenyl)ethoxy]-5-(1-piperidin-4-yl-1H-pyrazol-4-yl)pyridin-2-amine US10543199, Compound Crizotinib US9126941, PF-2341066 US11517561, Compound Crizotinib CRIZOTINIB 3-(2,6-dichloro-3-fluorobenzyloxy)-5-(1-(piperidin-4-yl)-1H-pyrazol-4-yl)pyridin-2-amine US10780082, Compound Crizotinib US9226923, Crizotinib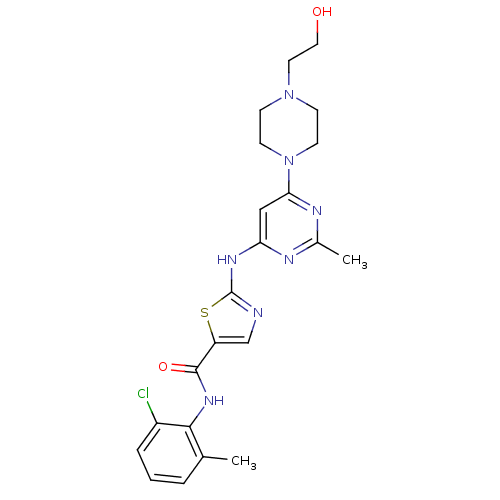 med.21724, Compound Dasatinib US20230348453, Compound A8 US10294227, Code Dasatinib BDBM13216 BMS-354825 CHEMBL1421 DASATINIB cid_3062316 N-(2-Chloro-6-methylphenyl)-2-[[6-[4-(2-hydroxyethyl)-1-piperazinyl)]-2-methyl-4-pyrimidinyl]amino)]-1,3-thiazole-5-carboxamide N-(2-chloro-6-methylphenyl)-2-({6-[4-(2-hydroxyethyl)piperazin-1-yl]-2-methylpyrimidin-4-yl}amino)-1,3-thiazole-5-carboxamide
med.21724, Compound Dasatinib US20230348453, Compound A8 US10294227, Code Dasatinib BDBM13216 BMS-354825 CHEMBL1421 DASATINIB cid_3062316 N-(2-Chloro-6-methylphenyl)-2-[[6-[4-(2-hydroxyethyl)-1-piperazinyl)]-2-methyl-4-pyrimidinyl]amino)]-1,3-thiazole-5-carboxamide N-(2-chloro-6-methylphenyl)-2-({6-[4-(2-hydroxyethyl)piperazin-1-yl]-2-methylpyrimidin-4-yl}amino)-1,3-thiazole-5-carboxamide
- Veach, DR; Namavari, M; Pillarsetty, N; Santos, EB; Beresten-Kochetkov, T; Lambek, C; Punzalan, BJ; Antczak, C; Smith-Jones, PM; Djaballah, H; Clarkson, B; Larson, SM Synthesis and biological evaluation of a fluorine-18 derivative of dasatinib. J Med Chem 50: 5853-7 (2007)
- Kwarcinski, FE; Brandvold, KR; Phadke, S; Beleh, OM; Johnson, TK; Meagher, JL; Seeliger, MA; Stuckey, JA; Soellner, MB Conformation-Selective Analogues of Dasatinib Reveal Insight into Kinase Inhibitor Binding and Selectivity. ACS Chem Biol 11: 1296-304 (2016)
- Li, HY; He, DD; Zhao, XJ; Sun, TY; Zhang, Q; Bai, CG; Chen, Y Design and synthesis of novel dasatinib derivatives as inhibitors of leukemia stem cells. Bioorg Med Chem Lett 28: 700-706 (2018)
- Hantschel, O; Rix, U; Schmidt, U; Bürckstümmer, T; Kneidinger, M; Schütze, G; Colinge, J; Bennett, KL; Ellmeier, W; Valent, P; Superti-Furga, G The Btk tyrosine kinase is a major target of the Bcr-Abl inhibitor dasatinib. Proc Natl Acad Sci U S A 104: 13283-8 (2007)
- Lim, JW; Kim, SK; Choi, SY; Kim, DH; Gadhe, CG; Lee, HN; Kim, HJ; Kim, J; Cho, SJ; Hwang, H; Seong, J; Jeong, KS; Lee, JY; Lim, SM; Lee, JW; Pae, AN Identification of crizotinib derivatives as potent SHIP2 inhibitors for the treatment of Alzheimer's disease. Eur J Med Chem 157: 405-422 (2018)
- Liu, L; Hussain, M; Luo, J; Duan, A; Chen, C; Tu, Z; Zhang, J Synthesis and biological evaluation of novel dasatinib analogues as potent DDR1 and DDR2 kinase inhibitors. Chem Biol Drug Des 89: 420-427 (2017)
- Liu, X; Zhang, L; Wan, H; Zhu, Z; Jin, J; Qin, Y; Mao, W; Yan, K; Fang, D; Jiang, W; Hu, L; Chen, J; Chen, K; Chen, S; Li, J; Zhao, S; Zheng, S; Zhang, L; Ding, CZ Discovery and preclinical evaluations of WX-0593, a novel ALK inhibitor targeting crizotinib-resistant mutations. Bioorg Med Chem Lett 66: (2022)
- Huang, Q; Johnson, TW; Bailey, S; Brooun, A; Bunker, KD; Burke, BJ; Collins, MR; Cook, AS; Cui, JJ; Dack, KN; Deal, JG; Deng, YL; Dinh, D; Engstrom, LD; He, M; Hoffman, J; Hoffman, RL; Johnson, PS; Kania, RS; Lam, H; Lam, JL; Le, PT; Li, Q; Lingardo, L; Liu, W; Lu, MW; McTigue, M; Palmer, CL; Richardson, PF; Sach, NW; Shen, H; Smeal, T; Smith, GL; Stewart, AE; Timofeevski, S; Tsaparikos, K; Wang, H; Zhu, H; Zhu, J; Zou, HY; Edwards, MP Design of potent and selective inhibitors to overcome clinical anaplastic lymphoma kinase mutations resistant to crizotinib. J Med Chem 57: 1170-87 (2014)
- Mathi, GR; Kang, CH; Lee, HK; Achary, R; Lee, HY; Lee, JY; Ha, JD; Ahn, S; Park, CH; Lee, CO; Hwang, JY; Yun, CS; Jung, HJ; Cho, SY; Kim, HR; Kim, P Replacing the terminal piperidine in ceritinib with aliphatic amines confers activities against crizotinib-resistant mutants including G1202R. Eur J Med Chem 126: 536-549 (2017)
- Chen, W; Guo, X; Zhang, C; Ke, D; Zhang, G; Yu, Y Discovery of 2-aminopyridines bearing a pyridone moiety as potent ALK inhibitors to overcome the crizotinib-resistant mutants. Eur J Med Chem 183: (2019)
- Wang, Y; Chen, S; Hu, G; Wang, J; Gou, W; Zuo, D; Gu, Y; Gong, P; Zhai, X Discovery of novel 2,4-diarylaminopyrimidine analogues as ALK and ROS1 dual inhibitors to overcome crizotinib-resistant mutants including G1202R. Eur J Med Chem 143: 123-136 (2018)
- PF-06463922 is a potent and selective next-generation ROS1/ALK inhibitor capable of blocking crizotinib-resistant ROS1 mutations.
- Liu, S; Jiang, Y; Yan, R; Li, Z; Wan, S; Zhang, T; Wu, X; Hou, J; Zhu, Z; Tian, Y; Zhang, J Design, synthesis and biological evaluations of 2-amino-4-(1-piperidine) pyridine derivatives as novel anti crizotinib-resistant ALK/ROS1 dual inhibitors. Eur J Med Chem 179: 358-375 (2019)
- Johnson, TW; Gallego, RA; Brooun, A; Gehlhaar, D; McTigue, M Reviving B-Factors: Retrospective Normalized B-Factor Analysis of c-ros Oncogene 1 Receptor Tyrosine Kinase and Anaplastic Lymphoma Kinase L1196M with Crizotinib and Lorlatinib. ACS Med Chem Lett 9: 878-883 (2018)
- Cui, JJ; Tran-Dubé, M; Shen, H; Nambu, M; Kung, PP; Pairish, M; Jia, L; Meng, J; Funk, L; Botrous, I; McTigue, M; Grodsky, N; Ryan, K; Padrique, E; Alton, G; Timofeevski, S; Yamazaki, S; Li, Q; Zou, H; Christensen, J; Mroczkowski, B; Bender, S; Kania, RS; Edwards, MP Structure based drug design of crizotinib (PF-02341066), a potent and selective dual inhibitor of mesenchymal-epithelial transition factor (c-MET) kinase and anaplastic lymphoma kinase (ALK). J Med Chem 54: 6342-63 (2011)
- Das, J; Chen, P; Norris, D; Padmanabha, R; Lin, J; Moquin, RV; Shen, Z; Cook, LS; Doweyko, AM; Pitt, S; Pang, S; Shen, DR; Fang, Q; de Fex, HF; McIntyre, KW; Shuster, DJ; Gillooly, KM; Behnia, K; Schieven, GL; Wityak, J; Barrish, JC 2-aminothiazole as a novel kinase inhibitor template. Structure-activity relationship studies toward the discovery of N-(2-chloro-6-methylphenyl)-2-[[6-[4-(2-hydroxyethyl)-1- piperazinyl)]-2-methyl-4-pyrimidinyl]amino)]-1,3-thiazole-5-carboxamide (dasatinib, BMS-354825) as a potent pan-Src kinase i J Med Chem 49: 6819-32 (2006)
- ChEMBL_2060935 (CHEMBL4716188) Inhibition of crizotinib-resistant ALK G1202R mutant (unknown origin) using peptide substrate incubated for 30 mins in presence of ATP by fluorescence assay
- ChEMBL_2060936 (CHEMBL4716189) Inhibition of crizotinib-resistant ALK L1196M mutant (unknown origin) using peptide substrate incubated for 30 mins in presence of ATP by fluorescence assay
- ChEMBL_2060937 (CHEMBL4716190) Inhibition of crizotinib-resistant ALK G1269A mutant (unknown origin) using peptide substrate incubated for 30 mins in presence of ATP by fluorescence assay
- ChEMBL_2060939 (CHEMBL4716192) Inhibition of crizotinib-resistant ALK F1174L mutant (unknown origin) using peptide substrate incubated for 30 mins in presence of ATP by fluorescence assay
- ChEMBL_2060940 (CHEMBL4716193) Inhibition of crizotinib-resistant ALK C1156Y mutant (unknown origin) using peptide substrate incubated for 30 mins in presence of ATP by fluorescence assay
- ChEMBL_2362669 Inhibition of wild type human BCR-ABL1 using Tyr2 peptide as substrate incubated for 1 hrs in presence of dasatinib by FRET based Z-LYTE assay
- Inhibition of SIK; Abl and Src Kinases by Other Kinase Inhibitor The IC50 for the inhibition of ABL1 by dasatinib, by C7 and by B3 is approximately 1.5 nM, 5.1 nM and 1.6 nM, and of SRC is 1.5 nM, 1.5 nM and 1.5 nM; each, respectively for dasatinib, C7 and by B3 (Table 3A). The compound C12 is also a strong inhibitor of SRC (<100 nM IC50), and is selective to SRC over ABL1. Briefly, a radiometric protein kinase assay (33PanQinase Activity Assay) was used for measuring the kinase activity of the five protein kinases. All kinase assays were performed in 96-well FlashPlates™ from PerkinElmer (Boston, MA, USA) in a 50 uL reaction volume. The reaction cocktail was pipetted in four steps in the following order:25 uL of assay buffer (standard buffer/[gamma-33P]-ATP)10 uL of ATP solution (in water)5 uL of test compound (in 10% DMSO)20 uL enzyme/substrate mix. The assay for all protein kinases contained 70 mM HEPES-NaOH pH7.5, 3 mM MgCl2, 3 mM MnCl2, 3 μM Na-orthovanadate, 1.2 mM DTT, ATP (variable concentrations, corresponding to the apparent ATP-Km of the respective kinase, see Table 2A), [gamma-33P]-ATP (approx. 8 105 cpm per well), protein kinase (variable amount, see Table 2A), and substrate (variable amounts, see Table 2A).
- End Point Fluorescence Assay Reaction volumes of 50 μL were used in 96-well plates. Buffer A (1×, 34 μL; 100 mM Tris, pH 8, 10 mM MgCl2) was added to a single row, followed by 15 μL ofenzyme (3.3× concentration) in buffer A with 3-fold dilutions (typically, 125 nM and 42, 14, 4.6, 1.5, 0.5, 0.17, 0.06, 0.02, 0.01, and 0 μM final well concentrations in buffer A). Then, 1 μL of a 500 nM stock of the appropriate dasatinib analogue BODIPY probe in DMSO was added (2% DMSO final). Wells were incubated at rt for 30 min prior to end point read (ex/em 485/535 nm). Reactions had final concentrations of 10 nM BODIPY-probe, 100 mM Tris buffer (pH 8),and 10 mM MgCl2.
- Inhibitory Activity of Compounds on TRKA and ALK-L1196M Based on HTRF of Cisbio (Cisbio, Cat. 08-52) principle, TRKA and ALK-L1196M kinase activity detection platform were established to determine the inhibitory activity of compounds. The compound powder was dissolved in 100% DMSO (Sigma, Cat. D8418-11) to prepare a 10 mM storage solution. The compounds had an initial test concentration of 1000 nM and 10,000 nM respectively, were 3-fold serially diluted to obtain 11 samples for multiple hole inspection. RXDX-101 (WuXi AppTec. supplied) or Crizotinib (WuXi AppTec. Supplied) was used as positive reference compound.The gradient diluted compounds were mixed with 0.5 nM TRKA (Carna, Cat. 08-186)/ALK-L1196M (Carna, Cat. 08-529), 0.3 μM/1 μM TK Substrate-biotin and 90 μM/30 μM ATP (Sigma, Cat. A7699) in a Optiplate-384F plate (PerkinElmer, Cat. 6007299) and incubated at room temperature for 90 mins/120 mins. Then 0.67 nM Eu-TK-Antibody and 50 nM Streptavidin-XL-665 were added, mixed and incubated at room temperature for 60 min. The fluorescence value was obtained by Envision (PerkinElmer, #2014) (Excitation light, 320 nm; Emission light, 665 nm). The IC50 values of the compounds were calculated using XLFIT5 (IDBS) software.
- Cell-Based Assays of c-Kit Mutant Kinase Activity The c-Kit mutant D816V inhibitors were assessed using an engineered BaF3-FL KIT D816V or BaF3-FL KIT V560G/D816V cell line. The BaF3-FL KIT D816V cell lines were created by introduction of KIT mutant (D816V) full length constructs that render the cells dependent on the introduced kinase for growth. Inhibitors of c-Kit mutant D816V kinase reduce or eliminate the mediated c-kit mutant D816V kinase activation, resulting in reduced cell proliferation of the BaF3-FL Kit mutant D816V cells. This inhibition is measured by the effect of compound concentration on cell growth to assess IC50 values. BaF3-FL KIT D816V cells were seeded at 1×104 cells per well of a 96 well cell culture plate in 50 μl of cell culture medium of RPMI Medium 1× (Invitrogen #11875-093) supplemented with 10% FBS (Invitrogen #10438), 1% Non Essential Amino Acids (Invitrogen #11140), 1% Penicillin Streptomycin (Invitrogen #15140), 1% L-Glutamine (Invitrogen #25030-081). Compounds were dissolved in DMSO at a concentration of 5 mM and were serially diluted 1:3 for a total of eight points and added to the cells to a final maximum concentration of 10 μM in 100 μl cell culture medium (final concentration 0.2% DMSO). Cells were also treated with Dasatinib as a positive control. The cells were incubated at 37° C., 5% CO2 for three days. ATPlite Buffer (Perkin Elmer #6016739) and substrate were equilibrated to room temperature, and enzyme/substrate Recombinant Firefly Luciferase/D-Luciferin was reconstituted. The cell plates were equilibrated to room temperature for 30 minutes, then lysed by addition of 25 uL per well of the ATPlite Reagent. The plate was mixed for 5 minutes on a plate shaker to lyse the cells. The plates were read on a Tecan Safire using Luminescence protocol modified to read 0.1s per well. The luminescence reading assesses the ATP content, which correlates directly with cell number such that the reading as a function of compound concentration is used to determine the IC50 value.
 DASATINIB BDBM31089
DASATINIB BDBM31089 Dasatinib BDBM82130
Dasatinib BDBM82130 US12129258, Example Crizotinib BDBM50306682 US10370379, Crizotinib (R)-3-(1-(2,6-dichloro-3-fluorophenyl)ethoxy)-5-(1-(piperidin-4-yl)-1H-pyrazol-4-yl)pyridin-2-amine CHEMBL601719 US11059827, Compound Crizotinib US9199944, Crizotinib PF-2341066 3-[(1R)-1-(2,6-dichloro-3-fluorophenyl)ethoxy]-5-(1-piperidin-4-yl-1H-pyrazol-4-yl)pyridin-2-amine US10543199, Compound Crizotinib US9126941, PF-2341066 US11517561, Compound Crizotinib CRIZOTINIB 3-(2,6-dichloro-3-fluorobenzyloxy)-5-(1-(piperidin-4-yl)-1H-pyrazol-4-yl)pyridin-2-amine US10780082, Compound Crizotinib US9226923, Crizotinib
US12129258, Example Crizotinib BDBM50306682 US10370379, Crizotinib (R)-3-(1-(2,6-dichloro-3-fluorophenyl)ethoxy)-5-(1-(piperidin-4-yl)-1H-pyrazol-4-yl)pyridin-2-amine CHEMBL601719 US11059827, Compound Crizotinib US9199944, Crizotinib PF-2341066 3-[(1R)-1-(2,6-dichloro-3-fluorophenyl)ethoxy]-5-(1-piperidin-4-yl-1H-pyrazol-4-yl)pyridin-2-amine US10543199, Compound Crizotinib US9126941, PF-2341066 US11517561, Compound Crizotinib CRIZOTINIB 3-(2,6-dichloro-3-fluorobenzyloxy)-5-(1-(piperidin-4-yl)-1H-pyrazol-4-yl)pyridin-2-amine US10780082, Compound Crizotinib US9226923, Crizotinib med.21724, Compound Dasatinib US20230348453, Compound A8 US10294227, Code Dasatinib BDBM13216 BMS-354825 CHEMBL1421 DASATINIB cid_3062316 N-(2-Chloro-6-methylphenyl)-2-[[6-[4-(2-hydroxyethyl)-1-piperazinyl)]-2-methyl-4-pyrimidinyl]amino)]-1,3-thiazole-5-carboxamide N-(2-chloro-6-methylphenyl)-2-({6-[4-(2-hydroxyethyl)piperazin-1-yl]-2-methylpyrimidin-4-yl}amino)-1,3-thiazole-5-carboxamide
med.21724, Compound Dasatinib US20230348453, Compound A8 US10294227, Code Dasatinib BDBM13216 BMS-354825 CHEMBL1421 DASATINIB cid_3062316 N-(2-Chloro-6-methylphenyl)-2-[[6-[4-(2-hydroxyethyl)-1-piperazinyl)]-2-methyl-4-pyrimidinyl]amino)]-1,3-thiazole-5-carboxamide N-(2-chloro-6-methylphenyl)-2-({6-[4-(2-hydroxyethyl)piperazin-1-yl]-2-methylpyrimidin-4-yl}amino)-1,3-thiazole-5-carboxamide