 GEFITINIB BDBM32367
GEFITINIB BDBM32367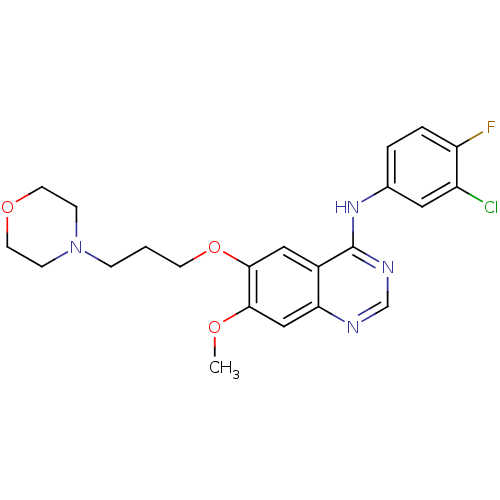 GEFITINIB Iressa N-(3-Chloro-4-fluorophenyl)-7-methoxy-6-[3-(4-morpholinyl)propoxy]-4-quinazolinamine BDBM5447 WO2022090481, Example gefitinib N-(3-chloro-4-fluorophenyl)-7-methoxy-6-[3-(morpholin-4-yl)propoxy]quinazolin-4-amine US9783524, Gefitinib US10507209, Compound Gefitinib US9416123, Gefitinib cid_123631 US10106508, Gefitinib CHEMBL939 ZD1839 US9730934, Gefitinib
GEFITINIB Iressa N-(3-Chloro-4-fluorophenyl)-7-methoxy-6-[3-(4-morpholinyl)propoxy]-4-quinazolinamine BDBM5447 WO2022090481, Example gefitinib N-(3-chloro-4-fluorophenyl)-7-methoxy-6-[3-(morpholin-4-yl)propoxy]quinazolin-4-amine US9783524, Gefitinib US10507209, Compound Gefitinib US9416123, Gefitinib cid_123631 US10106508, Gefitinib CHEMBL939 ZD1839 US9730934, Gefitinib US11524945, Compound Gefitinib BDBM582528
US11524945, Compound Gefitinib BDBM582528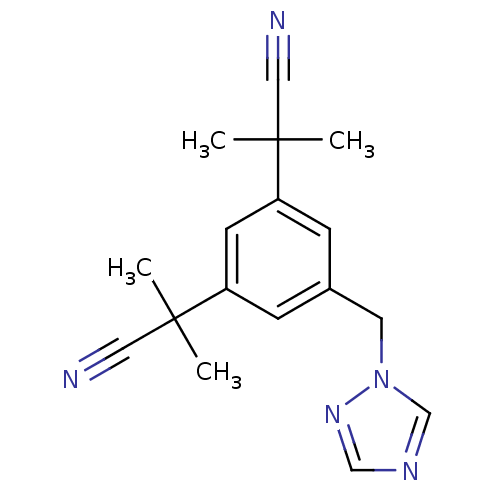 arimidex BDBM10015 NSC719344 2-[3-(1-cyano-1-methylethyl)-5-(1H-1,2,4-triazol-1-ylmethyl)phenyl]-2-methylpropanenitrile CHEMBL1399 2-[3-(2-cyanopropan-2-yl)-5-(1,2,4-triazol-1-ylmethyl)phenyl]-2-methyl-propanenitrile ANASTROZOLE
arimidex BDBM10015 NSC719344 2-[3-(1-cyano-1-methylethyl)-5-(1H-1,2,4-triazol-1-ylmethyl)phenyl]-2-methylpropanenitrile CHEMBL1399 2-[3-(2-cyanopropan-2-yl)-5-(1,2,4-triazol-1-ylmethyl)phenyl]-2-methyl-propanenitrile ANASTROZOLE BDBM50338602 Iressa N-(3-chloro-4-fluorophenyl)-7-methoxy-6-(3-morpholinopropoxy)quinazolin-4-amine 3-CHLORO-4-FLUORO-N-[(4Z)-7-METHOXY-6-(3-MORPHOLIN-4-YLPROPOXY)QUINAZOLIN-4(1H)-YLIDENE]ANILINE gefitinib ZD-1839 (3-Chloro-4-fluoro-phenyl)-[7-methoxy-6-(3-morpholin-4-yl-propoxy)-quinazolin-4-yl]-amine gifitinib (3-Chloro-4-fluoro-phenyl)-[7-methoxy-6-(2-morpholin-4-yl-ethoxy)-quinazolin-4-yl]-amine CHEMBL939 gefitnib
BDBM50338602 Iressa N-(3-chloro-4-fluorophenyl)-7-methoxy-6-(3-morpholinopropoxy)quinazolin-4-amine 3-CHLORO-4-FLUORO-N-[(4Z)-7-METHOXY-6-(3-MORPHOLIN-4-YLPROPOXY)QUINAZOLIN-4(1H)-YLIDENE]ANILINE gefitinib ZD-1839 (3-Chloro-4-fluoro-phenyl)-[7-methoxy-6-(3-morpholin-4-yl-propoxy)-quinazolin-4-yl]-amine gifitinib (3-Chloro-4-fluoro-phenyl)-[7-methoxy-6-(2-morpholin-4-yl-ethoxy)-quinazolin-4-yl]-amine CHEMBL939 gefitnib
- Edavana, VK; Dhakal, IB; Williams, S; Penney, R; Boysen, G; Yao-Borengasser, A; Kadlubar, S Potential role of UGT1A4 promoter SNPs in anastrozole pharmacogenomics. Drug Metab Dispos 41: 870-7 (2013)
- Wu, X; Li, M; Qu, Y; Tang, W; Zheng, Y; Lian, J; Ji, M; Xu, L Design and synthesis of novel Gefitinib analogues with improved anti-tumor activity. Bioorg Med Chem 18: 3812-22 (2010)
- Sun, M; Zhao, J; Chen, X; Zong, Z; Han, J; Du, Y; Sun, H; Wang, F Synthesis and biological evaluation of novel tricyclic oxazine and oxazepine fused quinazolines. Part 2: Gefitinib analogs. Bioorg Med Chem Lett 26: 4842-4845 (2016)
- Lin, H; Han, H; Yang, M; Wen, Z; Chen, Q; Ma, Y; Wang, X; Wang, C; Yin, T; Wang, X; Lu, G; Chen, H; Qi, J; Yang, Y PKM2/PDK1 dual-targeted shikonin derivatives restore the sensitivity of EGFR-mutated NSCLC cells to gefitinib by remodeling glucose metabolism. Eur J Med Chem 249: (2023)
- Engelman, JA; Zejnullahu, K; Gale, CM; Lifshits, E; Gonzales, AJ; Shimamura, T; Zhao, F; Vincent, PW; Naumov, GN; Bradner, JE; Althaus, IW; Gandhi, L; Shapiro, GI; Nelson, JM; Heymach, JV; Meyerson, M; Wong, KK; Jänne, PA PF00299804, an irreversible pan-ERBB inhibitor, is effective in lung cancer models with EGFR and ERBB2 mutations that are resistant to gefitinib. Cancer Res 67: 11924-32
- Song, Z; Huang, S; Yu, H; Jiang, Y; Wang, C; Meng, Q; Shu, X; Sun, H; Liu, K; Li, Y; Ma, X Synthesis and biological evaluation of morpholine-substituted diphenylpyrimidine derivatives (Mor-DPPYs) as potent EGFR T790M inhibitors with improved activity toward the gefitinib-resistant non-small cell lung cancers (NSCLC). Eur J Med Chem 133: 329-339 (2017)
- ChEMBL_1505545 (CHEMBL3595696) Inhibition of ADAM17 in human A549 cells assessed as potentiation of gefitinib-mediated inhibition of cell viability by measuring gefitinib IC50 at 40 uM incubated for 1 hr followed by gefitinib addition measured after 72 hrs by CellTiter-Glo assay (Rvb = 8.4 microM)
- ChEMBL_1487589 (CHEMBL3531895) Drug metabolism assessed as hecogenin-mediated inhibition of human recombinant UGT1A4-catalyzed anastrozole glucuronidation after 90 mins
- ChEMBL_1505615 (CHEMBL3595911) Inhibition of ADAM17 in human A549 cells assessed as potentiation of gefitinib-mediated inhibition of cell viability by measuring gefitinib IC50 at 10 uM after 96 hrs by CellTiter-Glo assay (Rvb = 8 microM)
- ChEMBL_2334390 Induction of GPX4 degradation in human Gefitinib-resistant H1650 cells incubated for 24 hrs by Western blot analysis
- ChEMBL_1922313 (CHEMBL4425269) Inhibition of gefitinib-resistant human GST-tagged EGFR L858R/T790M double mutant using pEY (4:1)/biotinylated pEY as substrate after 30 mins by ELISA
- Biochemical Assay Thus, while these compounds were extensively used in studying ILK-mediated cellular and disease processes, their reported inhibitory effects are probably due to unknown artifacts or indirect binding events. Next, we turned our attention to previously reported studies on kinase profiling and quantitative chemical proteomics. These studies suggested that a widely known lung cancer drug erlotinib, which targets EGFR, might also bind to ILK as an off target. By performing a robust fluorescence-based binding assay, we found that the FDA approved drug Erlotinib (TARCEVA) indeed binds potently to purified recombinant ILK at KD 0.43M, which is very close to the affinity of Erlotinib to EGFR measured at the same experimental conditions (KD 0.31 μM). Another erlotinib-like EGFR inhibitor Gefitinib exhibited 10-fold weaker binding affinity to ILK (KD 4.51 μM) yet 3-fold stronger affinity to EGFR (KD 0.11 μM) than erlotinib.
- V9103X ADP-Glo Kinase Assay Custom In a 384-well plate, test compound diluted in assay buffer (1% DMSO final) is mixed with 8His-RIPK2 FL enzyme (final concentration of 8 nM). After 15 minutes of pre-incubation at RT, ATP dissolved in assay buffer is added (final concentration 5 μM). The mixture is incubated for 60 minutes at 37° C. in a humidified incubator. Then, ADP Glo Reagent is added, followed by a 40 minute incubation at rt. Finally, Kinase Detection Reagent is added and the entire mixture is incubated for 40 min at RT. The luminescence signal is measured with an Envision reader to determine the amount of ADP produced. Assay buffer: 25 mM HEPES (4-(2-hydroxyethyl)-1-piperazinethanesulfonic acid), 0.1% BSA (bovine serum albumin), 10 mM MgCl2, 5 mM MnCl2, 50 mM KCl, 0.01% CHAPS (3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate), 10 μM Na3VO4, 1 mM DTT (dithiothreitol), pH 7.5 All plates contain wells with vehicle controls instead of compound (1% DMSO) as reference for the high signal (100% CTL (100% of control), high signal), and wells without enzyme as reference for low signal (0% CTL, low signal). The luminescent signal generated is proportional to the ADP produced and is correlated with enzyme activity. The analysis of the data is performed by the calculation of the percentage of ADP production in the presence of the test compound and RIPK2 as compared to the ADP production in the presence of RIPK2 plus 50 μM Gefitinib.
- Inhibition Assay Method: In a 384-well plate, test compound diluted in assay buffer (1% DMSO final) is mixed with 8His-RIPK2 FL enzyme (final concentration of 8 nM). After 15 minutes of pre-incubation at RT, ATP dissolved in assay buffer is added (final concentration 5 μM). The mixture is incubated for 60 minutes at 37° C. in a humidified incubator. Then, ADP Glo Reagent is added, followed by a 40 minute incubation at rt. Finally, Kinase Detection Reagent is added and the entire mixture is incubated for 40 min at RT. The luminescence signal is measured with an Envision reader to determine the amount of ADP produced. Assay buffer: 25 mM HEPES (4-(2-hydroxyethyl)-1-piperazinethanesulfonic acid), 0.1% BSA (bovine serum albumin), 10 mM MgCl2, 5 mM MnCl2, 50 mM KCl, 0.01% CHAPS (3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate), 10 μM Na3VO4, 1 mM DTT (dithiothreitol), pH 7.5 All plates contain wells with vehicle controls instead of compound (1% DMSO) as reference for the high signal (100% CTL (100% of control), high signal), and wells without enzyme as reference for low signal (0% CTL, low signal). The luminescent signal generated is proportional to the ADP produced and is correlated with enzyme activity. The analysis of the data is performed by the calculation of the percentage of ADP production in the presence of the test compound and RIPK2 as compared to the ADP production in the presence of RIPK2 plus 50 μM Gefitinib. (RLU (relative luminescence units)(sample)−RLU(low control))*100/(RLU(high value)−RLU(low control)) [RLU=relative luminescence units].
- High Throughput EGFR Biochemical Assay EGFR activity was measured using KinEASE (Cisbio), a time-resolved fluorescence resonance energy transfer (TR-FRET) immunoassay. In this assay, EGFR-catalyzes the phosphorylation of a universal Tyrosine kinase peptide substrate labeled with XL665. Europium conjugated phosphor-tyrosine specific antibody binds the resulting phosphorylated peptide. Formation of phosphorylated peptide is quantified by TR-FRET with Europium as the donor and XL665 the acceptor. The assay was performed in two main steps. The first step is the kinase reaction step and the second step is the detection step with TR-FRET reagents. In brief, test compounds 1:3 serially diluted in DMSO were delivered into Corning white, low volume, non-binding 384 well plates using the Echo 550 acoustic liquid dispenser (Labcyte ). EGFR enzyme (Human EGFR, cytoplasmic domain [669-1210] from Carna Biosciences Cat. No. 08-115) and substrates TK substrate-biotin (included in Cisbio HTRF KinEASE-TK kit Cat. No. 62TK0PEJ) were dispensed into assay plates using a Multi-Flo (Bio-Tek Instruments). The standard 10 μL reaction mixture contained 6 μM ATP (1×Km) or 12 μM ATP (2×Km), 1 μM biotinylated peptide, 0.3 nM EGFR (for 1×Km ATP) or 0.1 nM EGFR (for 2×Km ATP) in reaction buffer (10 mM MOPS, pH 7.0, 1.5% Glycerol, 0.5 mg/ml BSA, 10 mM Mg-Acetate, 1 mM DTT, 0.025% NP-40). After 60 min of incubation at room temperature, 10 μL of Stop and Detect Solution (1:400 Europium Cryptate labeled anti-phosphorylated peptide antibody solution and 125 nM strepavidin-XL665 Tracer in a 50 mM Hepes pH 7.0 detection buffer containing sufficient EDTA) was added. The plate was then further incubated for over 60 minutes at room temperature and read using an Envision 2103 Multilabeled reader (PerkinElmer) with excitation/emission/FRET emission at 340 nm/615 nm/665 nm, respectively. Fluorescence intensities at 615 nm and 665 nm emission wavelengths were expressed as a ratio (665 nm/615 nm). Percent inhibition was calculated as follows: % Inhibition=100x(RatioSample−Ratio0% Inhibition)/(Ratio100% Inhibition−Ratio0% Inhibition) where 0.05% DMSO (0% inhibition) was the negative control and 100 μM Staurosporine and Gefitinib (100% inhibition) was used as the positive control.
- High Throughput EGFR Biochemical Assay EGFR activity was measured using KinEASE (Cisbio), a time-resolved fluorescence resonance energy transfer (TR-FRET) immunoassay. In this assay, EGFR-catalyzes the phosphorylation of a universal Tyrosine kinase peptide substrate labeled with XL665. Europium conjugated phosphor-tyrosine specific antibody binds the resulting phosphorylated peptide. Formation of phosphorylated peptide is quantified by TR-FRET with Europium as the donor and XL665 the acceptor. The assay was performed in two main steps. The first step is the kinase reaction step and the second step is the detection step with TR-FRET reagents. In brief, test compounds 1:3 serially diluted in DMSO were delivered into Corning white, low volume, non-binding 384 well plates using the Echo 550 acoustic liquid dispenser (Labcyte ). EGFR enzyme (Human EGFR, cytoplasmic domain [669-1210] from Carna Biosciences Cat. No. 08-115) and substrates TK substrate-biotin (included in Cisbio HTRF KinEASE-TK kit Cat. No. 62TK0PEJ) were dispensed into assay plates using a Multi-Flo (Bio-Tek Instruments). The standard 10 μL reaction mixture contained 6 μM ATP (1×Km) or 12 μM ATP (2×Km), 1 μM biotinylated peptide, 0.3 nM EGFR (for 1×Km ATP) or 0.1 nM EGFR (for 2×Km ATP) in reaction buffer (10 mM MOPS, pH 7.0, 1.5% Glycerol, 0.5 mg/ml BSA, 10 mM Mg-Acetate, 1 mM DTT, 0.025% NP-40). After 60 min of incubation at room temperature, 10 μL of Stop and Detect Solution (1:400 Europium Cryptate labeled anti-phosphorylated peptide antibody solution and 125 nM strepavidin-XL665 Tracer in a 50 mM Hepes pH 7.0 detection buffer containing sufficient EDTA) was added. The plate was then further incubated for over 60 minutes at room temperature and read using an Envision 2103 Multilabeled reader (PerkinElmer) with excitation/emission/FRET emission at 340 nm/615 nm/665 nm, respectively. Fluorescence intensities at 615 nm and 665 nm emission wavelengths were expressed as a ratio (665 nm/615 nm). Percent inhibition was calculated as follows:% Inhibition=100×(RatioSample−Ratio0% Inhibition)/(Ratio100% Inhibition−Ratio0% Inhibition)where 0.05% DMSO (0% inhibition) was the negative control and 100 μM Staurosporine and Gefitinib (100% inhibition) was used as the positive control.
- In Vitro Enzyme Inhibitory Activity Samples:Controls: Gefitinib, erlotinib hydrochloride, purchased from Anqing worldchem Co., LTD.; lapatinib ditosylate, purchased from Taizhou Xingcheng Chempharm Co., Ltd.; CI-1033 hydrochloride, purchased from Shanghai hanxiangchem, Co., Ltd.; andThe present compounds: lab-made, their chemical names and structural formulae are shown in the preparation examples.Assay Procedures:The abbreviations used in the following assay have the following meanings:HEPES: hydroxyethyl piperazine ethanesulfonic acid;Brij-35: polyoxyethylene lauryl ether;DTT: dithiothreitol;Coating Reagent #3: #3 coating agent;EDTA: ethylene diamine tetraacetic acid, purchased from Sigma Co. Ltd.;FAM labeled peptide: fluorescein labeled peptide 22 (GL Biochem);ATP: adenosine triphosphate (Sigma);DMSO: dimethyl sulfoxide;EGFR: human epidermal growth factor receptor (Carna);HER2: human epidermal growth factor receptor 2 (Carna);HER4: human epidermal growth factor receptor 4 (Carna).1. Formulating the agents to be used in the assay(1) 1.25-fold MnCl2-free kinase buffer (62.5 mM HEPES, PH 7.5, 0.001875% Brij-35, 12.5 mM MgCl2, 2.5 mM DTT);(2) 1.25-fold MnCl2-containing kinase buffer (62.5 mM HEPES, pH 7.5, 0.001875% Brij-35, 12.5 mM MgCl2, 12.5 mM MnCl2, 2.5 mM DTT);(3) Stop buffer (100 mM HEPES, pH 7.5, 0.015% Brij-35, 0.2% Coating Reagent #3, 50 mM EDTA);(4) 2.5-fold kinase solutions (to the 1.25-fold kinase buffers were added the corresponding kinases to formulate 2.5-fold EGFR, HER2, HER4 kinase solutions);(5) 2.5-fold peptide solutions (to the 1.25-fold kinase buffers were added FAM labeled peptide and ATP to formulate the peptide solutions);(6) 5-fold compound solutions (using 100% DMSO to formulate 50-fold compound solutions having different concentration gradients, and diluting with water by 10 times to obtain 5-fold compound solutions having different concentration gradients);2. Adding 5 μL of a 5-fold compound solution to a 384-well plate;3. Adding 10 μL of a 2.5-fold kinase solution to incubate for 10 min;4. Then adding 10 μL of a 2.5-fold peptide solution, and reacting at 28° C. for 1 h; and5. Finally, adding 25 μL of stop buffer to terminate the reaction, and reading the data with Caliper.6. Curve fitting to obtain an IC50 value.The calculated inhibition ratio (%)=(the maximum conversion rate−the conversion rate)/(the maximum conversion rate−the minimum conversion rate)×100The curve fitting was conducted with the Xlfit software to obtain IC50 values.
 GEFITINIB BDBM32367
GEFITINIB BDBM32367 GEFITINIB Iressa N-(3-Chloro-4-fluorophenyl)-7-methoxy-6-[3-(4-morpholinyl)propoxy]-4-quinazolinamine BDBM5447 WO2022090481, Example gefitinib N-(3-chloro-4-fluorophenyl)-7-methoxy-6-[3-(morpholin-4-yl)propoxy]quinazolin-4-amine US9783524, Gefitinib US10507209, Compound Gefitinib US9416123, Gefitinib cid_123631 US10106508, Gefitinib CHEMBL939 ZD1839 US9730934, Gefitinib
GEFITINIB Iressa N-(3-Chloro-4-fluorophenyl)-7-methoxy-6-[3-(4-morpholinyl)propoxy]-4-quinazolinamine BDBM5447 WO2022090481, Example gefitinib N-(3-chloro-4-fluorophenyl)-7-methoxy-6-[3-(morpholin-4-yl)propoxy]quinazolin-4-amine US9783524, Gefitinib US10507209, Compound Gefitinib US9416123, Gefitinib cid_123631 US10106508, Gefitinib CHEMBL939 ZD1839 US9730934, Gefitinib US11524945, Compound Gefitinib BDBM582528
US11524945, Compound Gefitinib BDBM582528 arimidex BDBM10015 NSC719344 2-[3-(1-cyano-1-methylethyl)-5-(1H-1,2,4-triazol-1-ylmethyl)phenyl]-2-methylpropanenitrile CHEMBL1399 2-[3-(2-cyanopropan-2-yl)-5-(1,2,4-triazol-1-ylmethyl)phenyl]-2-methyl-propanenitrile ANASTROZOLE
arimidex BDBM10015 NSC719344 2-[3-(1-cyano-1-methylethyl)-5-(1H-1,2,4-triazol-1-ylmethyl)phenyl]-2-methylpropanenitrile CHEMBL1399 2-[3-(2-cyanopropan-2-yl)-5-(1,2,4-triazol-1-ylmethyl)phenyl]-2-methyl-propanenitrile ANASTROZOLE BDBM50338602 Iressa N-(3-chloro-4-fluorophenyl)-7-methoxy-6-(3-morpholinopropoxy)quinazolin-4-amine 3-CHLORO-4-FLUORO-N-[(4Z)-7-METHOXY-6-(3-MORPHOLIN-4-YLPROPOXY)QUINAZOLIN-4(1H)-YLIDENE]ANILINE gefitinib ZD-1839 (3-Chloro-4-fluoro-phenyl)-[7-methoxy-6-(3-morpholin-4-yl-propoxy)-quinazolin-4-yl]-amine gifitinib (3-Chloro-4-fluoro-phenyl)-[7-methoxy-6-(2-morpholin-4-yl-ethoxy)-quinazolin-4-yl]-amine CHEMBL939 gefitnib
BDBM50338602 Iressa N-(3-chloro-4-fluorophenyl)-7-methoxy-6-(3-morpholinopropoxy)quinazolin-4-amine 3-CHLORO-4-FLUORO-N-[(4Z)-7-METHOXY-6-(3-MORPHOLIN-4-YLPROPOXY)QUINAZOLIN-4(1H)-YLIDENE]ANILINE gefitinib ZD-1839 (3-Chloro-4-fluoro-phenyl)-[7-methoxy-6-(3-morpholin-4-yl-propoxy)-quinazolin-4-yl]-amine gifitinib (3-Chloro-4-fluoro-phenyl)-[7-methoxy-6-(2-morpholin-4-yl-ethoxy)-quinazolin-4-yl]-amine CHEMBL939 gefitnib