Compound (3)
Article Title (14)
Article Author (15)
Assay (25)
Suzuki, T; Kouketsu, A; Matsuura, A; Kohara, A; Ninomiya, S; Kohda, K; Miyata, N Thiol-based SAHA analogues as potent histone deacetylase inhibitors. Bioorg Med Chem Lett 14: 3313 -7 (2004) Desai, D; Salli, U; Vrana, KE; Amin, S SelSA, selenium analogs of SAHA as potent histone deacetylase inhibitors. Bioorg Med Chem Lett 20: 2044 -7 (2010) Tian, Y; Lv, W; Li, X; Wang, C; Wang, D; Wang, PG; Jin, J; Shen, J Stabilizing HDAC11 with SAHA to assay slow-binding benzamide inhibitors. Bioorg Med Chem Lett 27: 2943 -2945 (2017) Nardella, F; Halby, L; Dobrescu, I; Viluma, J; Bon, C; Claes, A; Cadet-Daniel, V; Tafit, A; Roesch, C; Hammam, E; Erdmann, D; Mairet-Khedim, M; Peronet, R; Mecheri, S; Witkowski, B; Scherf, A; Arimondo, PB Procainamide-SAHA Fused Inhibitors of hHDAC6 Tackle Multidrug-Resistant Malaria Parasites. J Med Chem 64: 10403 -10417 (2021) Rossi, M; Martinengo, B; Diamanti, E; Salerno, A; Rizzardi, N; Fato, R; Bergamini, C; Souza de Oliveira, A; de Araújo Marques Ferreira, T; Andrade Holanda, C; Romeiro, LAS; Soeiro, MNC; Nunes, K; Ferreira de Almeida Fiuza, L; Meuser Batista, M; Fraga, CAM; E A Alkhalaf, H; Elmahallawy, EK; Ebiloma, GU; De Koning, HP; Vittorio, S; Vistoli, G; Blanquart, C; Bertrand, P; Bolognesi, ML Benign-by-Design SAHA Analogues for Human and Animal Vector-Borne Parasitic Diseases. ACS Med Chem Lett 15: 1506 -1515 Bieliauskas, AV; Weerasinghe, SV; Pflum, MK Structural requirements of HDAC inhibitors: SAHA analogs functionalized adjacent to the hydroxamic acid. Bioorg Med Chem Lett 17: 2216 -9 (2007) Zhang, X; Zhang, J; Tong, L; Luo, Y; Su, M; Zang, Y; Li, J; Lu, W; Chen, Y The discovery of colchicine-SAHA hybrids as a new class of antitumor agents. Bioorg Med Chem 21: 3240 -4 (2013) Hendricks, JA; Keliher, EJ; Marinelli, B; Reiner, T; Weissleder, R; Mazitschek, R In vivo PET imaging of histone deacetylases by 18F-suberoylanilide hydroxamic acid (18F-SAHA). J Med Chem 54: 5576 -82 (2011) Negmeldin, AT; Padige, G; Bieliauskas, AV; Pflum, MK Structural Requirements of HDAC Inhibitors: SAHA Analogues Modified at the C2 Position Display HDAC6/8 Selectivity. ACS Med Chem Lett 8: 281 -286 (2017) Negmeldin, AT; Knoff, JR; Pflum, MKH The structural requirements of histone deacetylase inhibitors: C4-modified SAHA analogs display dual HDAC6/HDAC8 selectivity. Eur J Med Chem 143: 1790 -1806 (2018) Suzuki, T; Nagano, Y; Matsuura, A; Kohara, A; Ninomiya, S; Kohda, K; Miyata, N Novel histone deacetylase inhibitors: design, synthesis, enzyme inhibition, and binding mode study of SAHA-based non-hydroxamates. Bioorg Med Chem Lett 13: 4321 -6 (2003) Hanessian, S; Auzzas, L; Giannini, G; Marzi, M; Cabri, W; Barbarino, M; Vesci, L; Pisano, C Omega-alkoxy analogues of SAHA (vorinostat) as inhibitors of HDAC: a study of chain-length and stereochemical dependence. Bioorg Med Chem Lett 17: 6261 -5 (2007) Negmeldin, AT; Pflum, MKH The structural requirements of histone deacetylase inhibitors: SAHA analogs modified at the C5 position display dual HDAC6/8 selectivity. Bioorg Med Chem Lett 27: 3254 -3258 (2017) Suzuki, T; Nagano, Y; Kouketsu, A; Matsuura, A; Maruyama, S; Kurotaki, M; Nakagawa, H; Miyata, N Novel inhibitors of human histone deacetylases: design, synthesis, enzyme inhibition, and cancer cell growth inhibition of SAHA-based non-hydroxamates. J Med Chem 48: 1019 -32 (2005)
Adhikari, N; Halder, AK; Mallick, S; Saha, A; Saha, KD; Jha, T Bioorg Med Chem 24: 4291 -4309 (2016) Saha, AK; Yu, X; Lin, J; Lobera, M; Sharadendu, A; Chereku, S; Schutz, N; Segal, D; Marantz, Y; McCauley, D; Middleton, S; Siu, J; Börli, RW; Buys, J; Horner, M; Salyers, K; Schrag, M; Vargas, HM; Xu, Y; McElvain, M ACS Med Chem Lett 2: 97 -101 (2011) Chauhan, M; Saxena, A; Saha, B Eur J Med Chem 218: (2021) Saha, D; Ryan, KR; Lakkaniga, NR; Acharya, B; Garcia, NG; Smith, EL; Frett, B J Med Chem 64: 11747 -11773 (2021) Gillespie, P; Goodnow, RA; Saha, G; Bose, G; Moulik, K; Zwingelstein, C; Myers, M; Conde-Knape, K; Pietranico-Cole, S; So, SS Bioorg Med Chem Lett 24: 949 -53 (2014) Cifuentes-Pagano, E; Saha, J; Csányi, G; Ghouleh, IA; Sahoo, S; Rodríguez, A; Wipf, P; Pagano, PJ; Skoda, EM Medchemcomm 4: 1085 -1092 (2013) Milius, RA; Saha, JK; Madras, BK; Neumeyer, JL J Med Chem 34: 1728 -31 (1991) Adhikari, N; Halder, AK; Mallick, S; Saha, A; Saha, KD; Jha, T Bioorg Med Chem 24: 4291 -4309 (2016) Robello, M; Zheng, H; Saha, M; George Rosenker, KM; Debnath, S; Kumar, JP; Tagad, HD; Mazur, SJ; Appella, E; Appella, DH Eur J Med Chem 243: (2022) Kumar, R; Saha, N; Purohit, P; Garg, SK; Seth, K; Meena, VS; Dubey, S; Dave, K; Goyal, R; Sharma, SS; Banerjee, UC; Chakraborti, AK Eur J Med Chem 182: (2019) Zhang, H; Hsu, HC; Kahne, SC; Hara, R; Zhan, W; Jiang, X; Burns-Huang, K; Ouellette, T; Imaeda, T; Okamoto, R; Kawasaki, M; Michino, M; Wong, TT; Toita, A; Yukawa, T; Moraca, F; Vendome, J; Saha, P; Sato, K; Aso, K; Ginn, J; Meinke, PT; Foley, M; Nathan, CF; Darwin, KH; Li, H; Lin, G J Med Chem 64: 6262 -6272 (2021) Akhter, M; Akhter, N; Alam, MM; Zaman, MS; Saha, R; Kumar, A J Enzyme Inhib Med Chem 26: 767 -76 (2011) Balas, L; Durand, T; Saha, S; Johnson, I; Mukhopadhyay, S J Med Chem 52: 1005 -17 (2009) Makar, S; Saha, T; Singh, SK Eur J Med Chem 161: 252 -276 (2019) Kahraman, M; Sinishtaj, S; Dolan, PM; Kensler, TW; Peleg, S; Saha, U; Chuang, SS; Bernstein, G; Korczak, B; Posner, GH J Med Chem 47: 6854 -63 (2004)
HDAC6 fluorescence anisotropy assay The fluorescence anisotropy assay was performed with fluorescein-labeled SAHA (fl-SAHA, gift of C. Fierke, University of Michigan) and measured using a TECAN infinite F200Pro plate reader (λex = 485 nm, λem = 535 nm)60. Briefly, 30 nM fl-SAHA was titratedwith enzyme (0–15 μM) and incubated at room temperature for 20 min.The Kd values were determined by fitting the binding isotherm using GraphPad Prism software. ChEMBL_2307387 Inhibition of N-terminal nanoLuc-tagged YEATS3 in HEK293T cells co-transfected with C-terminal halo-tagged histone 3.3 incubated for 24 hrs in presence of SAHA by NanoBRET assay ChEMBL_2307400 Inhibition of N-terminal nanoLuc-tagged YEATS4 in HEK293T cells co-transfected with C-terminal halo-tagged histone 3.3 incubated for 24 hrs in presence of SAHA by NanoBRET assay ChEMBL_2076393 (CHEMBL4731927) Inhibition of HDAC6 (unknown origin) using BML-KI104 as substrate in presence of SAHA as inhibitor preincubated for 3 hrs followed by addition of substrate measured after 30 mins by fluorescence based assay ChEMBL_2076400 (CHEMBL4731934) Inhibition of class 1 HDAC in human Jurkat model of HIV latency assessed as reactivation of HIV latency incubated for 20 hrs in presence of SAHA and 5 % NHS by Steady-Glo luciferase assay ChEMBL_2076392 (CHEMBL4731926) Inhibition of human recombinant HDAC8 expressed in Escherichia coli using BML-KI178 as substrate and SAHA as inhibitor preincubated for 3 hrs followed by addition of substrate measured after 30 mins by fluorescence based assay ChEMBL_2081904 (CHEMBL4737695) Inhibition of Class 1 histone deacetylase in human THP-1 cells using Ac-LGK(Ac)-AMC as substrate incubated for 3 hrs followed by 50 uM SAHA addition and measured after 1 hr by fluorescence based assay ChEMBL_2076389 (CHEMBL4731923) Inhibition of human recombinant full length FLAG-tagged HDAC1 expressed in HEK293F cells using BML-KI104 as substrate in presence of SAHA as inhibitor preincubated for 3 hrs followed by addition of substrate measured after 30 mins by fluorescence based assay ChEMBL_2076390 (CHEMBL4731924) Inhibition of human recombinant full length FLAG-tagged HDAC2 expressed in baculovirus infected Sf9 cells using BML-KI104 as substrate in presence of SAHA as inhibitor preincubated for 3 hrs followed by addition of substrate measured after 30 mins by fluorescence based assay ChEMBL_2076391 (CHEMBL4731925) Inhibition of human recombinant full length FLAG-tagged HDAC3 expressed in HEK293F cells in using BML-KI104 as substrate in presence of SAHA as inhibitor preincubated for 3 hrs followed by addition of substrate measured after 30 mins by fluorescence based assay ChEMBL_2076394 (CHEMBL4731928) Inhibition of human recombinant full length HDAC11 expressed in baculovirus expression system using Boc-Lys(TFA)-AMC as substrate in presence of SAHA as inhibitor preincubated for 3 hrs followed by addition of substrate measured after 30 mins by fluorescence based assay ChEMBL_1672138 (CHEMBL4022167) Inhibition of recombinant C-terminal His-tagged HDAC11 (unknown origin) expressed in baculovirus infected Sf9 cells using Ac-Arg-Gly-Lys(Ac)-AMC as substrate pretreated for 3 hrs followed by substrate addition after 30 mins in presence of 0.2 uM SAHA by fluorometric method ChEMBL_2076398 (CHEMBL4731932) Inhibition of human recombinant full length N-terminal GST tagged HDAC5 expressed in sf9 insect cells using Boc-Lys(TFA)-AMC as substrate in presence of SAHA as inhibitor preincubated for 3 hrs followed by addition of substrate measured after 30 mins by fluorescence based assay HDAC8 Inhibition Assay (kinetic method) To determine the dissociation rate of the inhibitor, 1 µM HDAC8 and 20 µM inhibitor (syringe I) were mixed with 100 µM c-SAHA (syringe II) via the stopped-flow system. The fluorescence was measured at 325 nm with a 395 nm cutoff filter. Enzyme inhibition assay The nucleus extract was extracted from HDAC8 enzyme was expressed in Escherichia coli. Boc-Lys (acetyl)-AMC was used as the substrate of HDAC. SAHA which is the HDAC inhibitor on market was used as a positive control. The compounds were diluted to six concentrations (25, 5, 1, 0.2, 0.04 and 0.008 uM/L) to investigate their ability of inhibiting HDAC activity. Inhibitory Activity of the Compounds on HDAC6 Prepare a 10 mM stock solution of a compound with DMSO. Take 10 μl of stock solution and dilute with 90 μl DMSO to become a 1 mM working solution. The compound was three-fold serially diluted to total of 11 concentrations including a DMSO negative control. Add 3 μl of each concentration to 197 μl reaction buffer (20 mM Hepes pH 8.0, 137 mM NaCl, 2.7 mM KCl, 1 mM MgCl2, 0.05% BSA, 0.5 mM TCEP), mix well and add 10 μl to a 384-well plate. In the final reaction system, the concentrations of the compound were 10 μM to 0.51 nM. Duplicate, and add 10 μl 3XHDAC solution (BPS, Cat. 50006, 0.3 nM) to each well, followed by incubating at 23° C. for 20 minutes. Then, add 3× substrate solution (Anaspec, Cat. 61855, 15 μM), centrifuge to mix, and incubate at 23° C. for 90 minutes. Add 30 μl trypsin/SAHA mixture (20 mM Hepes pH 8.0, 100 mM NaCl, 10 mM SAHA, 0.01 mg/ml trypsin), incubate at 23° C. for 60 minutes to terminate the reaction. Finally, read fluorescence data by Envision (390 nm excitation, 460 nm emission). A high value at 430 nm indicates high kinase activity, while low value at 430 nm indicates that the kinase activity was inhibited. Finally, analyze the data by XLfit5 software and calculate the IC50 value of the compound. Vorinostat (SAHA) was a positive reference compound. HDAC2 Enzyme Assay The HDAC enzymatic assay was performed using a Fluorogenic HDAC Assay Kit (BPS Bioscience) according to the manufacturer's instructions. Briefly, HDAC2 enzymes were incubated with vehicle or various concentrations of the assayed samples or SAHA for 30 min at 37 °C in the presence of an HDAC fluorimetric substrate. The HDAC assay developer (which produces a fluorophore in reaction mixture) was added, and the fluorescence was measured using VICTOR3 (PerkinElmer, Waltham, MA, USA)with excitation at 360 nm and emission at 460 nm. HDAC6 Enzymatic Activity Assay (Alternative Protocol) Inhibition of HDAC enzymes was performed in 384-well plate format using human full-length recombinant HDAC1 and HDAC6, isolated from a baculovirus expression system in Sf9 cells (BPS Bioscience). Reaction buffer for HDAC1 contained 50 mM Tris HCl pH 8.0, 137 mM NaCl, 2.7 mM KCl, 1 mM MgCl2, 0.1 mg/mL BSA, and reaction buffer for HDAC6 contained 50 mM Tris/HCl, pH 8.0, 137 mM NaCl, 2.7 mM KCl, 250 μM EDTA, 1 mM DTT, 0.1 mg/mL BSA. Compounds (dissolved in 100% DMSO) or DMSO (vehicle) were diluted in assay buffer at 3× final assay concentration and then added to assay plates. SAHA (10 M) was used as a positive control. 3× final assay concentration of recombinant enzymes (final assay concentration is 4 nM and 5 nM for HDAC1 and HDAC6, respectively) were preincubated for 10 minutes with test compounds at room temperature. Afterwards, 3× final assay concentration of an acetylated fluorogenic peptide (Ac-Gly-Ala-Lys(Ac))-AMC, Bachem; final assay concentration is 12 M and 40 M for HDAC1 and HDAC6, respectively) was added to assay plate, allowing deacetylase reactions to incubate for 60 minutes at room temperature. The developer reagent containing 5 μM SAHA and 50 μM trypsin was added to stop the deacetylase reaction and generate AMNC-fluorescence. 15 minutes after addition of developer reagent, endpoint measurements were taken using CLARIOstar (BMG Labtech) plate reader (excitation/emission: 360/450). Fluorescence signals were normalized for each plate using GraphPad Prism software: reaction HDAC-substrate in presence of DMSO was set to 100% while reaction HDAC-substrate in presence of 10 μM SAHA was set to 0%. IC50-values were calculated from normalized measurements using GraphPad Prism software and nonlinear regression with 0% bottom and 100% top constraints. In vitro HDACs Inhibition Fluorescence Assay In brief, 10 μL of HeLa nuclear extract was mixed with various concentrations of target compounds (50 μL), SAHA, using 100% and none HDACs groups as control group, and the mixture. After incubation at 37 °C for 10 min, fluorogenic substrate Boc-Lys (acetyl)-AMC (40 μL) was added and then the mixture was incubated at 37 °C for 30 min. The mixture was stopped by addition of 100 μL of developer containing trypsin and TSA afterward. Over the next incubation at 37 °C for 20 min, fluorescence intensity was measured using a microplate reader at excitation and emission wavelengths of 390 and 460 nm, respectively. Reductase Activity Assay The HMGR activity was performed using HMG-CoA reductase assay kit from Sigma-Aldrich with the human recombinant protein or 100 μg total cell lysates from A549 cells. Lovastatin was used as a positive control, and SAHA as a negative control. HMGR activity under defined assay conditions, containing NADPH and HMG-CoA substrate in a final volume of 0.2 mL of 100 mM potassium phosphatate buffer (120 mM KCI, 1 mM EDTA, 5 mM DTT, pH 7.4), were initiated in the presence or absence (control) of test compounds dissolved in dimethylsulfoxide (DMSO). The rates of NADPH consumption were monitored every 20 seconds, for up to 10 minutes, by spectrophotometer at 37° C. and 340 nm. Reductase Activity Assay The HMGR activity was performed using HMG-CoA reductase assay kit from Sigma-Aldrich with the human recombinant protein or 100 μg total cell lysates from A549 cells. Lovastatin was used as a positive control, and SAHA as a negative control. HMGR activity under defined assay conditions, containing NADPH and HMG-CoA substrate in a final volume of 0.2 mL of 100 mM potassium phosphatate buffer (120 mM KCl, 1 mM EDTA, 5 mM DTT, pH 7.4), were initiated in the presence or absence (control) of test compounds dissolved in dimethylsulfoxide (DMSO). The rates of NADPH consumption were monitored every 20 seconds, for up to 10 minutes, by spectrophotometer at 37° C. and 340 nm. In vitro HDACs Inhibition Fluorescence Assay In brief, 10 μL of enzyme solution (HeLa nuclear extracts, HDAC1, HDAC4, HDAC6, HDAC8, HDAC11) was mixed with various concentrations of target compounds (50 μL), SAHA, and PXD101, using 100% and none HDACs groups as control group. Afterincubation at 37 °C for 10 min, 40 μL of fluorogenic substrate (Boc-Lys(Ac)-AMC for HeLa nuclear extracts; Ac-Leu- GlyLyS(Ac)-AMC for HDAC1, HDAC6, and HDAC11; Ac-Leu-GlyLyS(Tfa)-AMC for HDAC4 and HDAC8) was added, and then, the mixture was incubated at 37 °C for 30 min. The mixture was stopped by addition of 100 μL of developer containing trypsin and TSA afterward. Over the next incubation at 37 °C for 20 min, fluorescence intensity was measured using a microplate reader at excitation and emission wavelengths of 390 and 460 nm, respectively. Enzymatic Assay HDAC8 reagents: HDAC8 was purchased from Enzo Life Sciences [catalog #BML-SE145]. Assays were performed with buffer containing 20 mM HEPES, pH 8.0 [Boston BioProducts, catalog #BB-104, 1M stock], 100 mM NaCl [Sigma, catalog #S5150, 5M stock], 20 mM KCl [BioChemika, catalog #87526, 4M stock], 1 mM MgCl2 [Fluka, catalog #63020, 1M stock], 0.05% BSA (Fraction V) [Invitrogen, catalog #15260, 7.5% stock], and 0.1% n-Octyl-β-D-glucopyranoside (N-OG) [Anatrace, catalog #O311, 10% stock]. The HDAC8 enzyme was run at the final concentration of 1.333 nM. Fluor-de-Lys substrate [BioMol Research Laboratories, catalog #KI-178], used to evaluate enzyme activity, was added at the final concentration of 200 uM. To enable detection of the signal, Developer II [BioMol Research Laboratories, catalog #KI-176] was added at a 1:200 dilution to the stop solution, which also included 20 uM SAHA [Sigma, catalog #SML0061] to ensure complete termination of the reaction. Enzymatic Assay HDAC1 and 6 reagents: FLAG-tagged HDACs 1 and 6 were prepared in-house by protein expression in HEK293F cells followed by anti-FLAG affinity purification. Assays were performed with buffer containing 20 mM HEPES, pH 8.0 [Boston BioProducts, catalog #BB-104, 1M stock], 137 mM NaCl [Sigma, catalog #S5150, 5M stock], 2.7 mM KCl [BioChemika, catalog #87526, 4M stock], 1 mM MgCl2 [Fluka, catalog #63020, 1M stock], and 0.05% BSA (Fraction V) [Invitrogen, catalog #15260, 7.5% stock]. In addition to the above buffer ingredients, TCEP [CalBiochem, catalog #580561, 500 mM stock] was added at a final concentration of 0.5 mM to the buffer for the HDAC6 assays. HDAC1, 2, 3, and 6 enzymes were run at the final concentrations of 0.3 nM, 1.5 nM, 0.3 nM, and 1.333 nM, respectively. Fluor-de-Lys substrate [BioMol Research Laboratories, catalog #KI-104], used to evaluate enzyme activity, was added at the final concentrations of 20 uM, 40 uM, 20 uM, and 2.5 uM for HDACs 1, 2, 3, and 6. To enable detection of the signal, Developer [BioMol Research Laboratories, catalog #KI-105] was added at a 1:250 dilution to the stop solution, which also included 10 uM SAHA [Sigma, catalog #SML0061] to ensure complete termination of the reaction. Inhibitor Competition Assay Following transfection of HEK293 cells with pBJ5-HDAC1 wild type or mutant plasmids, [Weerasinghe et al., J. Med. Chem. 51:5542-5551; Wambua et al., J. Med. Chem. 57:642-650] as described above, cells were grown for 48 h and then subsequently treated with SAHA (10 uM in growth media containing DMEM, 10% FBS, 1% antibiotic/antimycotic, and <2% DMSO) for another 24 h before harvesting. Cells (20 x 1^06) were lysed in lysis buffer (500 uL; 50 mM Tris-Cl at pH 8.0, 150 mM NaCl, 10% glycerol, and 0.5% triton-X100) containing 1x protease inhibitor cocktail (GenDEPOT) at 4 °C for 30 min with rotation. The supernatant was collected using centrifugation at 13.2 x 10^3 rpm for 10 min at 4 °C. Prior to immunoprecipitation, anti-FLAG agarose beads (20 uL bead slurry) were washed with cold TBS (tris buffered saline; 20 mM Tris-Cl at pH 8.0, 150 mM NaCl) two times with spinning at 5000 rcf for 1 min at 4 °C. Wild type or mutant HDAC proteins were immunoprecipitated using the prewashed anti-FLAG agarose beads by incubating at 4 °C overnight with rotation. For inhibitor competition experiments, SHI-1:2 (10 uM in lysis buffer) or tubastatin (10 uM in lysis buffer) was included during immunoprecipitation. After immunoprecipitation, beads were washed three times with lysis buffer (1 mL), and bound proteins were eluted by incubating for 30 min at 4 °C using 3x FLAG peptide (APEXBIO; 50 uL; 0.25 mg mL in TBS). The eluted proteins were mixed with 4x SDS loading dye (25 uL; 100 mM Tris-Cl at pH 6.8, 4% SDS, 20% glycerol, 0.008% bromophenol blue, and 10% v/v β-mercaptoethanol), separated by 10% SDS-PAGE, and visualized with Sypro Ruby total protein stain (Molecular Probes) according to the manufacturer's instructions.
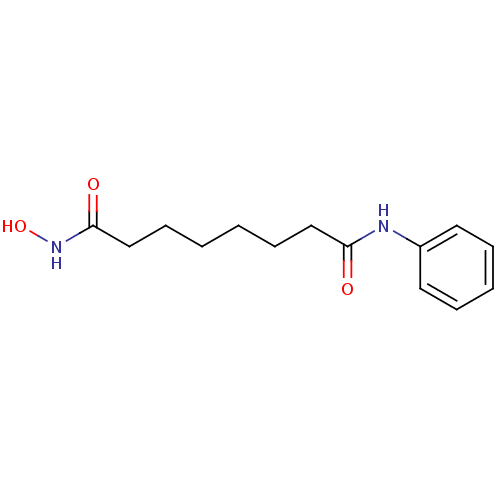 SAHA US11505523, Compound SAHA US9353061, SAHA BDBM19149 cid_5311 suberoylanilide hydroxamic acid N-hydroxy-N'-phenyloctanediamide US10011611, SAHA US9428447, SAHA US9695181, Vorinostat US10188756, Compound SAHA US11207431, SAHA Vorinostat US20240327418, Example SAHA US9115116, SAHA CHEMBL98 Zolinza
SAHA US11505523, Compound SAHA US9353061, SAHA BDBM19149 cid_5311 suberoylanilide hydroxamic acid N-hydroxy-N'-phenyloctanediamide US10011611, SAHA US9428447, SAHA US9695181, Vorinostat US10188756, Compound SAHA US11207431, SAHA Vorinostat US20240327418, Example SAHA US9115116, SAHA CHEMBL98 Zolinza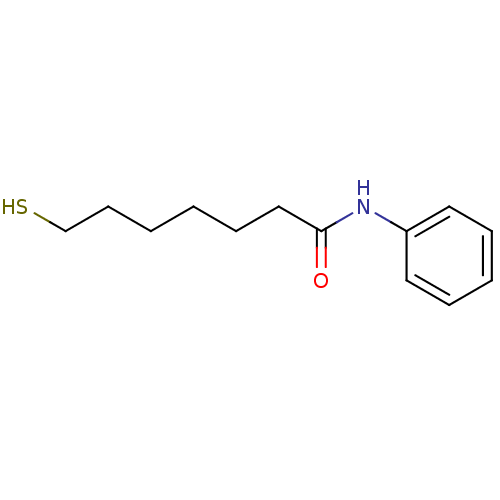 Thiol-SAHA (t-SAHA) BDBM152692
Thiol-SAHA (t-SAHA) BDBM152692 BDBM198125 fl-SAHA
BDBM198125 fl-SAHA