Query String: disulfiram DSF
 BDBM616976 US11753371, Compound Disulfiram
BDBM616976 US11753371, Compound Disulfiram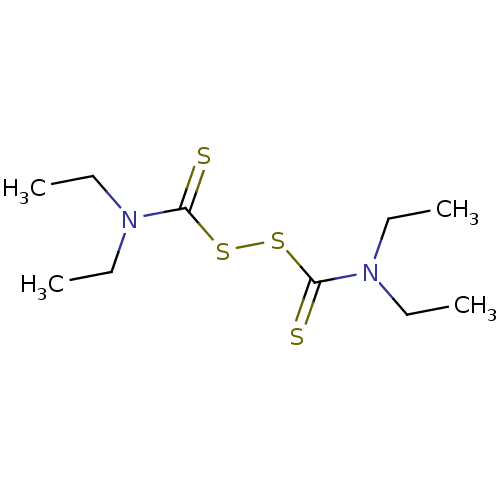 N,N,N',N'-tetraethylthiuram disulfide N,N-diethyl[(diethylcarbamothioyl)disulfanyl]carbothioamide (G5) BDBM50058655 US20230414581, Compound 37 Antabuse (TN) tetraethylthioperoxydicarbonic diamide DISULFIRAM med.21724, Compound 151 1,1',1'',1'''-[disulfanediylbis(carbonothioylnitrilo)]tetraethane Disul-firam bis(diethylthiocarbamoyl) disulfide CHEMBL964 tetraethylthiuram disulfide US20250145565, Compound Disulfiram cid_3117 1,1'-dithiobis(N,N-diethylthioformamide) tetraethylthiuram disulphide US11753371, Compound II-2a-(Disulfiram)
N,N,N',N'-tetraethylthiuram disulfide N,N-diethyl[(diethylcarbamothioyl)disulfanyl]carbothioamide (G5) BDBM50058655 US20230414581, Compound 37 Antabuse (TN) tetraethylthioperoxydicarbonic diamide DISULFIRAM med.21724, Compound 151 1,1',1'',1'''-[disulfanediylbis(carbonothioylnitrilo)]tetraethane Disul-firam bis(diethylthiocarbamoyl) disulfide CHEMBL964 tetraethylthiuram disulfide US20250145565, Compound Disulfiram cid_3117 1,1'-dithiobis(N,N-diethylthioformamide) tetraethylthiuram disulphide US11753371, Compound II-2a-(Disulfiram)
- OMRAN, Z; ABDULLAH, O DITHIENYL DISULFIRAM DERIVATIVES AS SELECTIVE ALDH1A1 INHIBITORS US Patent US20250145565 (2025)
- Omran, Z Disulfiram derivatives as ALDH1A1 and MAGL inhibitors US Patent US11753371 (2023)
- Cvek, B The Promiscuity of Disulfiram in Medicinal Research. ACS Med Chem Lett 14: 1610-1614 (2023)
- Omran, Z Development of new disulfiram analogues as ALDH1a1-selective inhibitors. Bioorg Med Chem Lett 40: (2021)
- Conway, TT; DeMaster, EG; Goon, DJ; Shirota, FN; Nagasawa, HT Diethylcarbamoylating/nitroxylating agents as dual action inhibitors of aldehyde dehydrogenase: a disulfiram-cyanamide merger. J Med Chem 42: 4016-20 (1999)
- Huang, Y; Xu, Y; Song, R; Ni, S; Liu, J; Xu, Y; Ren, Y; Rao, L; Wang, Y; Wei, L; Feng, L; Su, C; Peng, C; Li, J; Wan, J Identification of the New Covalent Allosteric Binding Site of Fructose-1,6-bisphosphatase with Disulfiram Derivatives toward Glucose Reduction. J Med Chem 63: 6238-6247 (2020)
- ChEMBL_1812133 (CHEMBL4311593) Binding affinity to recombinant human CA1 by DSF-based fluorescence thermal shift assay
- ChEMBL_1812134 (CHEMBL4311594) Binding affinity to recombinant human CA2 by DSF-based fluorescence thermal shift assay
- ChEMBL_1812136 (CHEMBL4311596) Binding affinity to recombinant human CA4 by DSF-based fluorescence thermal shift assay
- ChEMBL_1812139 (CHEMBL4311599) Binding affinity to recombinant human CA6 by DSF-based fluorescence thermal shift assay
- ChEMBL_1812140 (CHEMBL4311600) Binding affinity to recombinant human CA7 by DSF-based fluorescence thermal shift assay
- ChEMBL_1812141 (CHEMBL4311601) Binding affinity to recombinant human CA9 by DSF-based fluorescence thermal shift assay
- ChEMBL_1812142 (CHEMBL4311602) Binding affinity to recombinant human CA12 by DSF-based fluorescence thermal shift assay
- ChEMBL_1812143 (CHEMBL4311603) Binding affinity to recombinant human CA13 by DSF-based fluorescence thermal shift assay
- ChEMBL_2291844 Binding affinity to recombinant GSK3B (unknown origin) assessed as dissociation constant by DSF assay
- ChEMBL_2291845 Binding affinity to recombinant BMP2K (unknown origin) assessed as dissociation constant by DSF assay
- ChEMBL_2291846 Binding affinity to recombinant BMPR2 (unknown origin) assessed as dissociation constant by DSF assay
- ChEMBL_2291847 Binding affinity to recombinant FLT1 (unknown origin) assessed as dissociation constant by DSF assay
- ChEMBL_2291848 Binding affinity to recombinant MARK3 (unknown origin) assessed as dissociation constant by DSF assay
- ChEMBL_2291849 Binding affinity to recombinant MARK4 (unknown origin) assessed as dissociation constant by DSF assay
- ChEMBL_2291850 Binding affinity to recombinant MAPK15 (unknown origin) assessed as dissociation constant by DSF assay
- ChEMBL_2291851 Binding affinity to recombinant CDK2 (unknown origin) assessed as dissociation constant by DSF assay
- ChEMBL_2291852 Binding affinity to recombinant STK4 (unknown origin) assessed as dissociation constant by DSF assay
- ChEMBL_2291853 Binding affinity to recombinant MST3 (unknown origin) assessed as dissociation constant by DSF assay
- ChEMBL_2291854 Binding affinity to recombinant MERTK (unknown origin) assessed as dissociation constant by DSF assay
- ChEMBL_1707014 (CHEMBL4058247) Binding affinity myocilin-OLF domain (unknown origin) by Sypro Orange dye-based DSF assay
- ChEMBL_1812145 (CHEMBL4311605) Binding affinity to recombinant human CA1 assessed as intrinsic dissociation constant by DSF-based fluorescence thermal shift assay
- ChEMBL_1812146 (CHEMBL4311606) Binding affinity to recombinant human CA2 assessed as intrinsic dissociation constant by DSF-based fluorescence thermal shift assay
- ChEMBL_1812148 (CHEMBL4311608) Binding affinity to recombinant human CA4 assessed as intrinsic dissociation constant by DSF-based fluorescence thermal shift assay
- ChEMBL_1812151 (CHEMBL4311611) Binding affinity to recombinant human CA6 assessed as intrinsic dissociation constant by DSF-based fluorescence thermal shift assay
- ChEMBL_1812152 (CHEMBL4311612) Binding affinity to recombinant human CA7 assessed as intrinsic dissociation constant by DSF-based fluorescence thermal shift assay
- ChEMBL_1812153 (CHEMBL4311613) Binding affinity to recombinant human CA9 assessed as intrinsic dissociation constant by DSF-based fluorescence thermal shift assay
- ChEMBL_1812154 (CHEMBL4311614) Binding affinity to recombinant human CA12 assessed as intrinsic dissociation constant by DSF-based fluorescence thermal shift assay
- ChEMBL_1812155 (CHEMBL4311615) Binding affinity to recombinant human CA13 assessed as intrinsic dissociation constant by DSF-based fluorescence thermal shift assay
- ChEMBL_2184649 (CHEMBL5096731) Binding affinity to C-terminal domain human STING (139 to 379 residues) assessed as dissociation constant by DSF method
- ChEMBL_2426105 Binding affinity to human ClpP assessed as thermal stabilization incubated for 1 hr by SYPRO orange dye based DSF analysis
- ChEMBL_2465709 Binding affinity to human Casein lysing protease P (HsClpP) assessed as change in melting temperature measured after 2 hrs differential scanning fluorimetry (DSF) assay
- ChEMBL_1812137 (CHEMBL4311597) Binding affinity to recombinant full length C-terminal His6x-tagged human CA5A expressed in Escherichia coli BL21(DE3) by DSF-based fluorescence thermal shift assay
- ChEMBL_1812138 (CHEMBL4311598) Binding affinity to recombinant full length N-terminal His6x-tagged human CA5B expressed in Escherichia coli Rosetta 2 (DE3) by DSF-based fluorescence thermal shift assay
- ChEMBL_1812135 (CHEMBL4311595) Binding affinity to recombinant N-terminal His6x-tagged human CA3 (4 to 260 residues) expressed in Escherichia coli BL21(DE3) by DSF-based fluorescence thermal shift assay
- ChEMBL_1812144 (CHEMBL4311604) Binding affinity to recombinant N-terminal His6x-tagged human CA14 (20 to 280 residues) catalytic domain expressed in Escherichia coli Rosetta 2 (DE3) by DSF-based fluorescence thermal shift assay
- ChEMBL_1812149 (CHEMBL4311609) Binding affinity to recombinant full length C-terminal His6x-tagged human CA5A expressed in Escherichia coli BL21(DE3) assessed as intrinsic dissociation constant by DSF-based fluorescence thermal shift assay
- ChEMBL_1812150 (CHEMBL4311610) Binding affinity to recombinant full length N-terminal His6x-tagged human CA5B expressed in Escherichia coli Rosetta 2 (DE3) assessed as intrinsic dissociation constant by DSF-based fluorescence thermal shift assay
- ChEMBL_1812147 (CHEMBL4311607) Binding affinity to recombinant N-terminal His6x-tagged human CA3 (4 to 260 residues) expressed in Escherichia coli BL21(DE3) assessed as intrinsic dissociation constant by DSF-based fluorescence thermal shift assay
- ChEMBL_1812156 (CHEMBL4311616) Binding affinity to recombinant N-terminal His6x-tagged human CA14 (20 to 280 residues) catalytic domain expressed in Escherichia coli Rosetta 2 (DE3) assessed as intrinsic dissociation constant by DSF-based fluorescence thermal shift assay
- ChEMBL_2367757 Binding affinity to N-terminal His6-SUMO-tagged Mtb ClpP1 (7 to 200 residues)/ClpP2 (13 to 214 residues) complex expressed in Escherichia coli BL21(DE3) assessed as increase in thermal stability measured at 25 to 110 degreeC in presence of Bz-LL by DSF assay
- scanning fluorimetry assay of HPAEpiC cells The differential scanning fluorimetry (DSF) assay was carried out on a CFX96 real-time PCR Detection System (Bio-Rad). All the experiments were performed in the following buffer: 20 mM HEPES pH 6.5 containing 120 mM NaCl. The Mpro protein (final concentration: 2 μM) was pre-incubated with 40 μM of compounds for 30 min. 5×SYPRO Orange dye (Sigma) was added to probe the thermal denaturation from 20°C to 95°C at a scan rate of 1.5 °C/min. The melt temperature (Tm) was calculated by using a Boltzmann model in GraphPad Prism 8.0 software. The thermal shift (ΔTm) was calculated using the equation ΔTm=Tm(compound)-Tm(DMSO). All experiments were performed in triplicate, and the values are presented as mean ± SD. HPAEpiC cells are used.
- scanning fluorimetry assay of Vero E6 cells The differential scanning fluorimetry (DSF) assay was carried out on a CFX96 real-time PCR Detection System (Bio-Rad). All the experiments were performed in the following buffer: 20 mM HEPES pH 6.5 containing 120 mM NaCl. The Mpro protein (final concentration: 2 μM) was pre-incubated with 40 μM of compounds for 30 min. 5×SYPRO Orange dye (Sigma) was added to probe the thermal denaturation from 20°C to 95°C at a scan rate of 1.5 °C/min. The melt temperature (Tm) was calculated by using a Boltzmann model in GraphPad Prism 8.0 software. The thermal shift (ΔTm) was calculated using the equation ΔTm=Tm(compound)-Tm(DMSO). All experiments were performed in triplicate, and the values are presented as mean ± SD. Vero E6 cells are used.