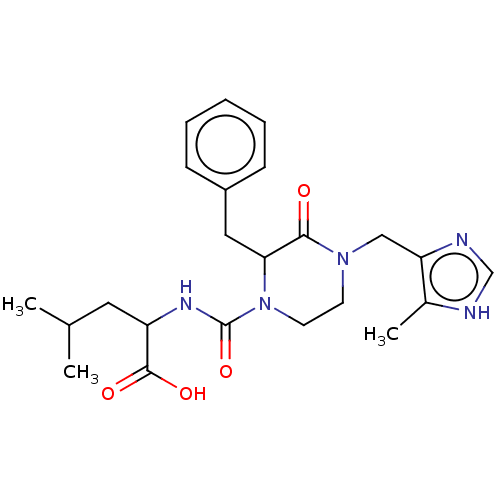
 US11078214, Compound PTX BDBM511066
US11078214, Compound PTX BDBM511066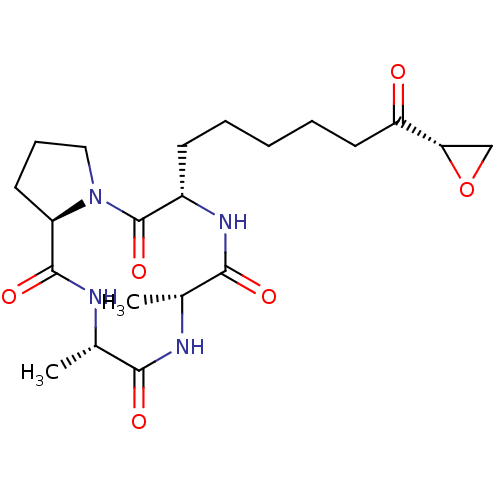 HC toxin BDBM198122
HC toxin BDBM198122 BDBM50161873 CHEMBL437624 E[c(RGDyK)]2-PTX conjugate
BDBM50161873 CHEMBL437624 E[c(RGDyK)]2-PTX conjugate SMR001566822 MLS002703014 T-2 TOXIN cid_529495 BDBM93500
SMR001566822 MLS002703014 T-2 TOXIN cid_529495 BDBM93500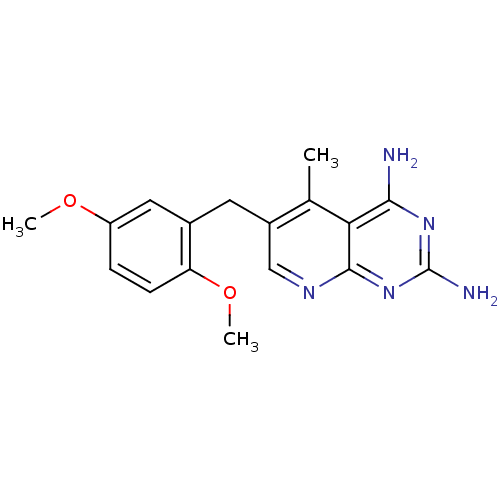 BDBM18224 6-[(2,5-dimethoxyphenyl)methyl]-5-methylpyrido[2,3-d]pyrimidine-2,4-diamine Piritrexim PTX [2-amino-6-(2,5-dimethoxybenzyl)-5-methyl-pyrido[2,3-d]pyrimidin-4-yl]amine;2-hydroxyethanesulfonic acid CHEMBL7492
BDBM18224 6-[(2,5-dimethoxyphenyl)methyl]-5-methylpyrido[2,3-d]pyrimidine-2,4-diamine Piritrexim PTX [2-amino-6-(2,5-dimethoxybenzyl)-5-methyl-pyrido[2,3-d]pyrimidin-4-yl]amine;2-hydroxyethanesulfonic acid CHEMBL7492 Joro spider toxin 3 CHEMBL313747 N*1*-(5-{3-[4-(3-Amino-propylamino)-butylamino]-propionylamino}-pentyl)-2-[2-(2,4-dihydroxy-phenyl)-acetylamino]-succinamide 2-Amino-N-{3-[4-(3-amino-propylamino)-butylamino]-propyl}-acetamide BDBM50105843
Joro spider toxin 3 CHEMBL313747 N*1*-(5-{3-[4-(3-Amino-propylamino)-butylamino]-propionylamino}-pentyl)-2-[2-(2,4-dihydroxy-phenyl)-acetylamino]-succinamide 2-Amino-N-{3-[4-(3-amino-propylamino)-butylamino]-propyl}-acetamide BDBM50105843
- Martin, L; Cornille, F; Turcaud, S; Meudal, H; Roques, BP; Fournié-Zaluski, MC Metallopeptidase inhibitors of tetanus toxin: A combinatorial approach. J Med Chem 42: 515-25 (1999)
- Williams, JD; Khan, AR; Cardinale, SC; Butler, MM; Bowlin, TL; Peet, NP Small molecule inhibitors of anthrax lethal factor toxin. Bioorg Med Chem 22: 419-34 (2013)
- Chen, D; Ma, L; Kanalas, JJ; Gao, J; Pawlik, J; Jimenez, ME; Walter, MA; Peterson, JW; Gilbertson, SR; Schein, CH Structure-based redesign of an edema toxin inhibitor. Bioorg Med Chem 20: 368-76 (2012)
- Saito, R; Pruet, JM; Manzano, LA; Jasheway, K; Monzingo, AF; Wiget, PA; Kamat, I; Anslyn, EV; Robertus, JD Peptide-conjugated pterins as inhibitors of ricin toxin A. J Med Chem 56: 320-9 (2013)
- Pan, BF; Sweet, DH; Pritchard, JB; Chen, R; Nelson, JA A transfected cell model for the renal toxin transporter, rOCT2. Toxicol Sci 47: 181-6 (1999)
- Sanchez, AM; Thomas, D; Gillespie, EJ; Damoiseaux, R; Rogers, J; Saxe, JP; Huang, J; Manchester, M; Bradley, KA Amiodarone and bepridil inhibit anthrax toxin entry into host cells. Antimicrob Agents Chemother 51: 2403-11 (2007)
- Reynaud, S; Laurin, SA; Ciolek, J; Barbe, P; Van Baelen, AC; Susset, M; Blondel, F; Ghazarian, M; Boeri, J; Vanden Driessche, M; Upert, G; Mourier, G; Kessler, P; Konnert, L; Beroud, R; Keck, M; Servent, D; Bouvier, M; Gilles, N From a Cone Snail Toxin to a Competitive MC4R Antagonist. J Med Chem 65: 12084-12094 (2022)
- Moayeri, M; Robinson, TM; Leppla, SH; Karginov, VA In vivo efficacy of beta-cyclodextrin derivatives against anthrax lethal toxin. Antimicrob Agents Chemother 52: 2239-41 (2008)
- Deguchi, T; Kusuhara, H; Takadate, A; Endou, H; Otagiri, M; Sugiyama, Y Characterization of uremic toxin transport by organic anion transporters in the kidney. Kidney Int 65: 162-74 (2004)
- Pruet, JM; Saito, R; Manzano, LA; Jasheway, KR; Wiget, PA; Kamat, I; Anslyn, EV; Robertus, JD Optimized 5-membered heterocycle-linked pterins for the inhibition of Ricin Toxin A. ACS Med Chem Lett 3: 588-591 (2012)
- Andersen, TF; Tikhonov, DB; Bølcho, U; Bolshakov, K; Nelson, JK; Pluteanu, F; Mellor, IR; Egebjerg, J; Strømgaard, K Uncompetitive antagonism of AMPA receptors: Mechanistic insights from studies of polyamine toxin derivatives. J Med Chem 49: 5414-23 (2006)
- Harvey, AJ; Gable, RW; Baell, JB A three-residue, continuous binding epitope peptidomimetic of ShK toxin as a Kv1.3 inhibitor. Bioorg Med Chem Lett 15: 3193-6 (2005)
- Lin, L; Olson, ME; Sugane, T; Turner, LD; Tararina, MA; Nielsen, AL; Kurbanov, EK; Pellett, S; Johnson, EA; Cohen, SM; Allen, KN; Janda, KD Catch and Anchor Approach To Combat Both Toxicity and Longevity of Botulinum Toxin A. J Med Chem 63: 11100-11120 (2020)
- Letourneau, JJ; Stroke, IL; Hilbert, DW; Sturzenbecker, LJ; Marinelli, BA; Quintero, JG; Sabalski, J; Ma, L; Diller, DJ; Stein, PD; Webb, ML Identification and initial optimization of inhibitors of Clostridium difficile (C. difficile) toxin B (TcdB). Bioorg Med Chem Lett 28: 756-761 (2018)
- Barslund, AF; Poulsen, MH; Bach, TB; Lucas, S; Kristensen, AS; Strømgaard, K Solid-phase synthesis and biological evaluation of Joro spider toxin-4 from Nephila clavata. J Nat Prod 74: 483-6 (2011)
- Jung, ME; Chamberlain, BT; Ho, CL; Gillespie, EJ; Bradley, KA Structure-Activity Relationship of Semicarbazone EGA Furnishes Photoaffinity Inhibitors of Anthrax Toxin Cellular Entry. ACS Med Chem Lett 5: 363-7 (2014)
- Miller, DJ; Ravikumar, K; Shen, H; Suh, JK; Kerwin, SM; Robertus, JD Structure-based design and characterization of novel platforms for ricin and shiga toxin inhibition. J Med Chem 45: 90-8 (2001)
- Mudd, GE; Scott, H; Chen, L; van Rietschoten, K; Ivanova-Berndt, G; Dzionek, K; Brown, A; Watcham, S; White, L; Park, PU; Jeffrey, P; Rigby, M; Beswick, P Discovery of BT8009: A Nectin-4 Targeting Bicycle Toxin Conjugate for the Treatment of Cancer. J Med Chem 65: 14337-14347 (2022)
- Gademann, K; Portmann, C; Blom, JF; Zeder, M; Jüttner, F Multiple toxin production in the cyanobacterium microcystis: isolation of the toxic protease inhibitor cyanopeptolin 1020. J Nat Prod 73: 980-4 (2010)
- Kurbanov, EK; Chiu, TL; Solberg, J; Francis, S; Maize, KM; Fernandez, J; Johnson, RL; Hawkinson, JE; Walters, MA; Finzel, BC; Amin, EA Probing the S2' Subsite of the Anthrax Toxin Lethal Factor Using Novel N-Alkylated Hydroxamates. J Med Chem 58: 8723-33 (2015)
- Cummer, R; Grosjean, F; Bolteau, R; Vasegh, SE; Veyron, S; Keogh, L; Trempe, JF; Castagner, B Structure-Activity Relationship of Inositol Thiophosphate Analogs as Allosteric Activators of Clostridioides difficile Toxin B. J Med Chem 67: 16576-16597
- Booth, RG; Trevor, A; Singer, TP; Castagnoli, N Studies on semirigid tricyclic analogues of the nigrostriatal toxin 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine. J Med Chem 32: 473-7 (1989)
- Gagnon, H; Beauchemin, S; Kwiatkowska, A; Couture, F; D'Anjou, F; Levesque, C; Dufour, F; Desbiens, AR; Vaillancourt, R; Bernard, S; Desjardins, R; Malouin, F; Dory, YL; Day, R Optimization of furin inhibitors to protect against the activation of influenza hemagglutinin H5 and Shiga toxin. J Med Chem 57: 29-41 (2014)
- Kromann, H; Krikstolaityte, S; Andersen, AJ; Andersen, K; Krogsgaard-Larsen, P; Jaroszewski, JW; Egebjerg, J; Strømgaard, K Solid-phase synthesis of polyamine toxin analogues: potent and selective antagonists of Ca2+-permeable AMPA receptors. J Med Chem 45: 5745-54 (2002)
- Chauhan, V; Chaudhary, D; Pathak, U; Saxena, N; Dhaked, RK In Silico Discovery and Validation of Amide Based Small Molecule Targeting the Enzymatic Site of Shiga Toxin. J Med Chem 59: 10763-10773 (2016)
- Letourneau, JJ; Stroke, IL; Hilbert, DW; Cole, AG; Sturzenbecker, LJ; Marinelli, BA; Quintero, JG; Sabalski, J; Li, Y; Ma, L; Pechik, I; Stein, PD; Webb, ML Synthesis and SAR studies of novel benzodiazepinedione-based inhibitors of Clostridium difficile (C. difficile) toxin B (TcdB). Bioorg Med Chem Lett 28: 3601-3605 (2018)
- Israel, MR; Thongyoo, P; Deuis, JR; Craik, DJ; Vetter, I; Durek, T The E15R Point Mutation in Scorpion Toxin Cn2 Uncouples Its Depressant and Excitatory Activities on Human Na J Med Chem 61: 1730-1736 (2018)
- Reynaud, S; Ciolek, J; Degueldre, M; Saez, NJ; Sequeira, AF; Duhoo, Y; Brás, JLA; Meudal, H; Cabo Díez, M; Fernández Pedrosa, V; Verdenaud, M; Boeri, J; Pereira Ramos, O; Ducancel, F; Vanden Driessche, M; Fourmy, R; Violette, A; Upert, G; Mourier, G; Beck-Sickinger, AG; Mörl, K; Landon, C; Fontes, CMGA; Miñambres Herráiz, R; Rodríguez de la Vega, RC; Peigneur, S; Tytgat, J; Quinton, L; De Pauw, E; Vincentelli, R; Servent, D; Gilles, N A Venomics Approach Coupled to High-Throughput Toxin Production Strategies Identifies the First Venom-Derived Melanocortin Receptor Agonists. J Med Chem 63: 8250-8264 (2020)
- Mayorga-Flores, M; Chantôme, A; Melchor-Meneses, CM; Domingo, I; Titaux-Delgado, GA; Galindo-Murillo, R; Vandier, C; Del Río-Portilla, F Novel Blocker of Onco SK3 Channels Derived from Scorpion Toxin Tamapin and Active against Migration of Cancer Cells. ACS Med Chem Lett 11: 1627-1633 (2020)
- Liu, J; Xu, S; Huang, C; Shen, J; Yu, S; Yu, Y; Sun, Q; Dai, Q Synthesis and activity evaluation of selenazole-coupled CPI-1 irreversible bifunctional inhibitors for botulinum toxin A light chain. Bioorg Med Chem Lett 73: (2022)
- Hoque, MA; Islam, MN; Islam, MS; Kato, T; Nishino, N; Ito, A; Yoshida, M Design and synthesis of CHAP31, trapoxin B and HC-toxin based bicyclic tetrapeptides disulfide as potent histone deacetylase inhibitors. Bioorg Med Chem 22: 3850-5 (2014)
- Wittwer, MB; Zur, AA; Khuri, N; Kido, Y; Kosaka, A; Zhang, X; Morrissey, KM; Sali, A; Huang, Y; Giacomini, KM Discovery of potent, selective multidrug and toxin extrusion transporter 1 (MATE1, SLC47A1) inhibitors through prescription drug profiling and computational modeling. J Med Chem 56: 781-95 (2013)
- PubChem, PC High-throughput multiplex microsphere dose response for inhibitors of toxin protease, specifically Lethal Factor protease, compounds from Cherry Pick 01 PubChem Bioassay (2012)
- PubChem, PC High-throughput multiplex microsphere dose response for inhibitors of toxin protease, specifically Lethal Factor protease, compounds from Cherry Pick 02 PubChem Bioassay (2012)
- PubChem, PC High-throughput multiplex microsphere dose response for inhibitors of toxin protease, specifically Lethal Factor protease, compounds from Powder Set 01 PubChem Bioassay (2012)
- O'Malley, S; Sareth, S; Jiao, GS; Kim, S; Thai, A; Cregar-Hernandez, L; McKasson, L; Margosiak, SA; Johnson, AT Virtual medicinal chemistry: in silico pre-docking functional group transformation for discovery of novel inhibitors of botulinum toxin serotype A light chain. Bioorg Med Chem Lett 23: 2505-11 (2013)
- PubChem, PC High-throughput multiplex microsphere dose response for inhibitors of toxin protease, specifically Botulinum neurotoxin light chain A protease, compounds from Cherry Pick 01 PubChem Bioassay (2012)
- PubChem, PC High-throughput multiplex microsphere dose response for inhibitors of toxin protease, specifically Botulinum neurotoxin light chain A protease, compounds from Cherry Pick 02 PubChem Bioassay (2012)
- PubChem, PC High-throughput multiplex microsphere dose response for inhibitors of toxin protease, specifically Botulinum neurotoxin light chain A protease, compounds from Powder Set 01 PubChem Bioassay (2012)
- PubChem, PC High-throughput multiplex microsphere dose response for inhibitors of toxin protease, specifically Botulinum neurotoxin light chain F protease, compounds from Cherry Pick 01 PubChem Bioassay (2012)
- PubChem, PC High-throughput multiplex microsphere dose response for inhibitors of toxin protease, specifically Botulinum neurotoxin light chain F protease, compounds from Cherry Pick 02 PubChem Bioassay (2012)
- PubChem, PC High-throughput multiplex microsphere dose response for inhibitors of toxin protease, specifically Botulinum neurotoxin light chain F protease, compounds from Powder Set 01 PubChem Bioassay (2012)
- Wang, S; Yang, J; Li, X; Liu, Z; Wu, Y; Si, G; Tao, Y; Zhao, N; Hu, X; Ma, Y; Liu, G Discovery of 1,4-Benzodiazepine-2,5-dione (BZD) Derivatives as Dual Nucleotide Binding Oligomerization Domain Containing 1/2 (NOD1/NOD2) Antagonists Sensitizing Paclitaxel (PTX) To Suppress Lewis Lung Carcinoma (LLC) Growth in Vivo. J Med Chem 60: 5162-5192 (2017)
- Hume, WE; Shingaki, T; Takashima, T; Hashizume, Y; Okauchi, T; Katayama, Y; Hayashinaka, E; Wada, Y; Kusuhara, H; Sugiyama, Y; Watanabe, Y The synthesis and biodistribution of [(11)C]metformin as a PET probe to study hepatobiliary transport mediated by the multi-drug and toxin extrusion transporter 1 (MATE1) in vivo. Bioorg Med Chem 21: 7584-90 (2013)
- ChEMBL_934992 (CHEMBL2320361) Inhibition of Ricin toxin A
- ChEMBL_1823892 (CHEMBL4323656) Inhibition of Clostridium difficile toxin B
- ChEMBL_2309303 Inhibition of Clostridium difficile full length Toxin B
- ChEMBL_609720 (CHEMBL1067457) Inhibition of Clostridium botulinum toxin BoNT/A light chain
- ChEMBL_1353638 (CHEMBL3271072) Inhibition of Bacillus anthracis lethal toxin-induced mouse RAW264.7 cell death preincubated for 1 hr followed by lethal toxin challenge measured after 4 hrs
- ChEMBL_609713 (CHEMBL1067450) Inhibition of Clostridium botulinum toxin BoNT/A light chain by FRET assay
- ChEMBL_934991 (CHEMBL2320360) Inhibition of Ricin toxin A after 90 mins by luciferase-translational assay
- ChEMBL_2147703 (CHEMBL5032049) Binding affinity to castor beans ricin toxin A subunit assessed as dissociation constant
- ChEMBL_609714 (CHEMBL1067451) Inhibition of Clostridium botulinum toxin BoNT/A light chain by HPLC-based assay
- ChEMBL_1282600 (CHEMBL3101399) Inhibition of castor bean ricin toxin A after 90 mins by luciferase reporter gene assay
- ChEMBL_1621659 (CHEMBL3863942) Binding affinity to recombinant Shigella dysenteriae type 1 Shiga toxin A subunit by SPR method
- ChEMBL_840713 (CHEMBL2090455) Inhibition of Ricinus communis ricin toxin A after 90 mins by luciferase reporter gene assay
- ChEMBL_2016866 (CHEMBL4670444) Inhibition of Clostridium botulinum BoNT/A intoxicated in hiPSC-derived neurons assessed as reduction in SNAP-25 cleavage using SNAP-25 as substrate treated 30 mins post toxin addition and measured 7 hrs post toxin addition by Western blot analysis
- ChEMBL_1564790 (CHEMBL3784393) Inhibition of NLRP3 in mouse J774 macrophages assessed as reduction in Bacillus anthracis lethal toxin induced cell death
- ChEMBL_2147706 (CHEMBL5032052) Binding affinity to castor beans ricin toxin A subunit assessed as dissociation constant by surface plasmon resonance analysis
- ChEMBL_884651 (CHEMBL2214667) Inhibition of Bacillus anthracis edema toxin-mediated cAMP production in mouse RAW264.7 cells after 4 hrs by ELISA
- ChEMBL_2147704 (CHEMBL5032050) Inhibition of castor beans ricin toxin A subunit assessed as depurination of ribosome using liver ribosomes by qRT-PCR analysis
- ChEMBL_2147708 (CHEMBL5032054) Inhibition of castor beans ricin toxin A subunit assessed as depurination of ribosome using yeast ribosomes by qRT-PCR analysis
- ChEMBL_1286970 (CHEMBL3111135) Inhibition of castor bean ricin-induced toxicity in human A549 cells assessed as protection against toxin-induced inhibition of protein biosynthesis by measuring incorporation of [14C]-leucine into cells preincubated for 1 hr followed by toxin challenge measured after 20 hrs by liquid scintillation counting analysis
- ChEBML_1696093 Agonist activity at human D2SR expressed in HEK293 cell membranes co-expressing PTX insensitive variant of Galphao1 incubated for 30 mins by [35S]GTP-gammaS binding assay
- ChEMBL_2487221 Allosteric activation of Clostridioides difficile Toxin B assessed as induction of autoproteolysis measured after 3 hrs by Western blot based extent cleavage assay
- ChEMBL_1696093 (CHEMBL4046983) Agonist activity at human D2SR expressed in HEK293 cell membranes co-expressing PTX insensitive variant of Galphao1 incubated for 30 mins by [35S]GTP-gammaS binding assay
- ChEMBL_554441 (CHEMBL953930) Protection against Bacillus anthracis lethal toxin-mediated cytotoxicity in mouse RAW264.7 cells assessed as change in viability after 24 hrs by WST1 dye reduction assay
- ChEMBL_1621651 (CHEMBL3863934) Inhibition of Shigella dysenteriae type 1 Shiga toxin A subunit assessed as change in transition temperature by SYPRO orange dye based fluorescence-based thermal shift assay
- ChEMBL_2330580 Negative allosteric modulator activity at P-gp in human LCC6MDR cells overexpressing P-gp assessed as reversal of P-gp mediated PTX resistance by measuring paclitaxel IC50 incubated for 5 days by MTS assay
- ChEMBL_2487222 Allosteric activation of Clostridioides difficile Toxin B assessed as induction of autoproteolysis measured after 3 hrs in presence of CaCl2, MgCl2, ZnCl2 by Western blot based extent cleavage assay
- ChEMBL_2487219 Binding affinity to His6 tagged Clostridioides difficile Toxin B truncated cysteine protease domain (543 to 799 residues) extracted from Escherichia coli BL21 (DE3) assessed as dissociation constant by ITC method
- ChEMBL_1752668 (CHEMBL4187428) Inhibition of Clostridium difficile toxin B transfected in CHO cells assessed as reduction in caspase 3/7 activation pre-incubated for 1 hr before TcdB addition by luminescence based assay1
- ChEMBL_1752669 (CHEMBL4187429) Inhibition of Clostridium difficile toxin A transfected in CHO cells assessed as reduction in caspase 3/7 activation pre-incubated for 1 hr before TcdA addition by luminescence based assay1
- ChEMBL_1828993 (CHEMBL4328867) Inhibition of Clostridium difficile toxin B transfected in CHO cells assessed as reduction in caspase 3/7 activation pre-incubated for 1 hr before TcdB addition by luminescence based assay
- ChEMBL_1828994 (CHEMBL4328868) Inhibition of Clostridium difficile toxin A transfected in CHO cells assessed as reduction in caspase 3/7 activation pre-incubated for 1 hr before TcdA addition by luminescence based assay
- ChEMBL_2330597 Negative allosteric modulator activity at human wildtype P-gp expressed in HEK293FT cells assessed as reversal of P-gp mediated PTX resistance by measuring paclitaxel IC50 incubated for 5 days by MTS assay (Rvb = 540.5 +/- 20.3 nM)
- ChEMBL_2330596 Negative allosteric modulator activity at human P-gp G1114A mutant expressed in HEK293FT cells assessed as reversal of P-gp mediated PTX resistance by measuring paclitaxel IC50 incubated for 5 days by MTS assay (Rvb = 5.6 +/- 1.1 nM)
- ChEMBL_2330598 Negative allosteric modulator activity at human P-gp I1115A mutant expressed in HEK293FT cells assessed as reversal of P-gp mediated PTX resistance by measuring paclitaxel IC50 incubated for 5 days by MTS assay (Rvb = 7.5 +/- 2.4 nM)
- ChEMBL_2330599 Negative allosteric modulator activity at human P-gp H1195A mutant expressed in HEK293FT cells assessed as reversal of P-gp mediated PTX resistance by measuring paclitaxel IC50 incubated for 5 days by MTS assay (Rvb = 3.4 +/- 1.5 nM)
- ChEMBL_2330600 Negative allosteric modulator activity at human P-gp T1226A mutant expressed in HEK293FT cells assessed as reversal of P-gp mediated PTX resistance by measuring paclitaxel IC50 incubated for 5 days by MTS assay (Rvb = 6.3 +/- 2.1 nM)
- ChEMBL_2330601 Negative allosteric modulator activity at human P-gp Q1193A mutant expressed in HEK293FT cells assessed as reversal of P-gp mediated PTX resistance by measuring paclitaxel IC50 incubated for 5 days by MTS assay (Rvb = 483.4 +/- 17.2 nM)
- ChEMBL_2330602 Negative allosteric modulator activity at human P-gp L1113A mutant expressed in HEK293FT cells assessed as reversal of P-gp mediated PTX resistance by measuring paclitaxel IC50 incubated for 5 days by MTS assay (Rvb = 590 +/- 14.2 nM)
- ChEMBL_2330603 Negative allosteric modulator activity at human P-gp C1227A mutant expressed in HEK293FT cells assessed as reversal of P-gp mediated PTX resistance by measuring paclitaxel IC50 incubated for 5 days by MTS assay (Rvb = 352.1 +/- 30.6 nM)
- ChEMBL_2330604 Negative allosteric modulator activity at human P-gp Q1193F mutant expressed in HEK293FT cells assessed as reversal of P-gp mediated PTX resistance by measuring paclitaxel IC50 incubated for 5 days by MTS assay (Rvb = 453.6 +/- 14.8 nM)
- ChEMBL_2330605 Negative allosteric modulator activity at human P-gp Q1193E mutant expressed in HEK293FT cells assessed as reversal of P-gp mediated PTX resistance by measuring paclitaxel IC50 incubated for 5 days by MTS assay (Rvb = 395.8 +/- 13.4 nM)
- ChEMBL_2330606 Negative allosteric modulator activity at human P-gp Q1193K mutant expressed in HEK293FT cells assessed as reversal of P-gp mediated PTX resistance by measuring paclitaxel IC50 incubated for 5 days by MTS assay (Rvb = 466.2 +/- 72.3 nM)
- ChEMBL_2330607 Negative allosteric modulator activity at human P-gp Q1193C mutant expressed in HEK293FT cells assessed as reversal of P-gp mediated PTX resistance by measuring paclitaxel IC50 incubated for 5 days by MTS assay (Rvb = 359.4 +/- 22.3 nM)
- ChEMBL_2330608 Negative allosteric modulator activity at human P-gp Q1193T mutant expressed in HEK293FT cells assessed as reversal of P-gp mediated PTX resistance by measuring paclitaxel IC50 incubated for 5 days by MTS assay (Rvb = 523.6 +/- 33.7 nM)
- ChEMBL_2330609 Negative allosteric modulator activity at human P-gp Q1193N mutant expressed in HEK293FT cells assessed as reversal of P-gp mediated PTX resistance by measuring paclitaxel IC50 incubated for 5 days by MTS assay (Rvb = 560.2 +/- 43.9 nM)
- ChEMBL_2330610 Negative allosteric modulator activity at human P-gp G1114V mutant expressed in HEK293FT cells assessed as reversal of P-gp mediated PTX resistance by measuring paclitaxel IC50 incubated for 5 days by MTS assay (Rvb = 3.4 +/- 0.5 nM)
- ChEMBL_2330611 Negative allosteric modulator activity at human P-gp G1114I mutant expressed in HEK293FT cells assessed as reversal of P-gp mediated PTX resistance by measuring paclitaxel IC50 incubated for 5 days by MTS assay (Rvb = 10.2 +/- 2.4 nM)
- ChEMBL_2330612 Negative allosteric modulator activity at human P-gp G1114L mutant expressed in HEK293FT cells assessed as reversal of P-gp mediated PTX resistance by measuring paclitaxel IC50 incubated for 5 days by MTS assay (Rvb = 4.5 +/- 1.1 nM)
- ChEMBL_2330613 Negative allosteric modulator activity at human P-gp I1115W mutant expressed in HEK293FT cells assessed as reversal of P-gp mediated PTX resistance by measuring paclitaxel IC50 incubated for 5 days by MTS assay (Rvb = 7.8 +/- 0.9 nM)
- ChEMBL_2330614 Negative allosteric modulator activity at human P-gp I1115Y mutant expressed in HEK293FT cells assessed as reversal of P-gp mediated PTX resistance by measuring paclitaxel IC50 incubated for 5 days by MTS assay (Rvb = 6.9 +/- 1.2 nM)
- ChEMBL_2330615 Negative allosteric modulator activity at human P-gp I1115K mutant expressed in HEK293FT cells assessed as reversal of P-gp mediated PTX resistance by measuring paclitaxel IC50 incubated for 5 days by MTS assay (Rvb = 6.2 +/- 2 nM)
- ChEMBL_2330616 Negative allosteric modulator activity at human P-gp I1115E mutant expressed in HEK293FT cells assessed as reversal of P-gp mediated PTX resistance by measuring paclitaxel IC50 incubated for 5 days by MTS assay (Rvb = 7.4 +/- 0.8 nM)
- ChEMBL_2330617 Negative allosteric modulator activity at human P-gp I1115N mutant expressed in HEK293FT cells assessed as reversal of P-gp mediated PTX resistance by measuring paclitaxel IC50 incubated for 5 days by MTS assay (Rvb = 10.2 +/- 1.6 nM)
- ChEMBL_2330618 Negative allosteric modulator activity at human P-gp I1115M mutant expressed in HEK293FT cells assessed as reversal of P-gp mediated PTX resistance by measuring paclitaxel IC50 incubated for 5 days by MTS assay (Rvb = 9.8 +/- 0.9 nM)
- ChEMBL_2330619 Negative allosteric modulator activity at human P-gp I1115F mutant expressed in HEK293FT cells assessed as reversal of P-gp mediated PTX resistance by measuring paclitaxel IC50 incubated for 5 days by MTS assay (Rvb = 663.1 +/- 10.2 nM)
- ChEMBL_2330620 Negative allosteric modulator activity at human P-gp I1115L mutant expressed in HEK293FT cells assessed as reversal of P-gp mediated PTX resistance by measuring paclitaxel IC50 incubated for 5 days by MTS assay (Rvb = 472 +/- 15.4 nM)
- ChEMBL_2330621 Negative allosteric modulator activity at human P-gp T1226W mutant expressed in HEK293FT cells assessed as reversal of P-gp mediated PTX resistance by measuring paclitaxel IC50 incubated for 5 days by MTS assay (Rvb = 363.5 +/- 24.9 nM)
- ChEMBL_2330622 Negative allosteric modulator activity at human P-gp T1226Y mutant expressed in HEK293FT cells assessed as reversal of P-gp mediated PTX resistance by measuring paclitaxel IC50 incubated for 5 days by MTS assay (Rvb = 449.3 +/- 31.2 nM)
- ChEMBL_2330623 Negative allosteric modulator activity at human P-gp T1226F mutant expressed in HEK293FT cells assessed as reversal of P-gp mediated PTX resistance by measuring paclitaxel IC50 incubated for 5 days by MTS assay (Rvb = 409.9 +/- 19.7 nM)
- ChEMBL_2487220 Binding affinity to His6 tagged Clostridioides difficile Toxin B truncated cysteine protease domain (543 to 799 residues) extracted from Escherichia coli BL21 (DE3) assessed as dissociation constant in presence of 10 mM Ca2+ by ITC method
- ChEMBL_1828995 (CHEMBL4328869) Inhibition of C-terminal 6-His tagged recombinant Clostridium difficile toxin B glucosyltransferase domain assessed as reduction in UDP-glucose hydrolysis activity using UDP-glucose substrate and ADP Alexa633 tracer incubated for 3 hrs by fluorescence polarization assay
- ChEMBL_855550 (CHEMBL2162855) Inhibition of Clostridium botulinum neurotoxin serotype A light chain in Sprague-Dawley rat cerebellar granule cells assessed as protection against SNAP-25 cleavage at pre-exposed to compound before toxin addition measured 4 hrs post dose by SDS-PAGE based densitometry
- ChEMBL_1621657 (CHEMBL3863940) Inhibition of Shigella dysenteriae type 1 Shiga toxin A subunit in African green monkey Vero cells assessed as inhibition of Stx-induced cytotoxicity pre-treated with compound for 1 hr followed by Stx exposure for 24 hrs by neutral red uptake assay
- ChEMBL_1752667 (CHEMBL4187427) Inhibition of C-terminal 6-His tagged recombinant Clostridium difficile toxin B catalytic fragment (Met1 to Leu543 residues) assessed as reduction in glucosyltransferase domain UDP-glucose hydrolysis activity using UDP-glucose and ADP Alexa633 tracer incubated fore 3 hrs by fluorescence polarization assay
- Dose response confirmation uHTS hits for MazEF TA System activators via a fluorescence-based single-stranded RNase assay Data Source: Sanford-Burnham Center for Chemical Genomics (SBCCG) Source Affiliation: Sanford-Burnham Medical Research Institute (SBMRI, San Diego, CA) Network: NIH Molecular Libraries Probe Production Centers Network (MLPCN) Grant Number: 2R01 GM068385-06 Assay Provider: Dr. Paul Hergenrother, University of Illinois, Urbana, IL Bacterial resistance to antibiotics is a worldwide health crisis. Resistance typically occurs as a result of chromosomal mutation or acquisition of a mobile genetic element, such as a plasmid, that harbors resistance-mediating genes. One of the most common plasmid maintenance systems is the toxin-antitoxin (TA) postsegregational killing mechanism. In this mechanism, if a plasmid-free daughter cell arises, the labile antitoxin is degraded and the toxin induces cell death. TA genes are ubiquitous in clinical isolates of certain drug-resistant bacteria, and it has been postulated that compounds that disrupt the TA interaction could free the toxin to kill the bac
- SAR analysis of small molecule activators of the MazEF TA System via a fluorescence-based single-stranded RNase assay Data Source: Sanford-Burnham Center for Chemical Genomics (SBCCG) Source Affiliation: Sanford-Burnham Medical Research Institute (SBMRI, San Diego, CA) Network: NIH Molecular Libraries Probe Production Centers Network (MLPCN) Grant Number: 2R01 GM068385-06 Assay Provider: Dr. Paul Hergenrother, University of Illinois, Urbana, IL Bacterial resistance to antibiotics is a worldwide health crisis. Resistance typically occurs as a result of chromosomal mutation or acquisition of a mobile genetic element, such as a plasmid, that harbors resistance-mediating genes. One of the most common plasmid maintenance systems is the toxin-antitoxin (TA) postsegregational killing mechanism. In this mechanism, if a plasmid-free daughter cell arises, the labile antitoxin is degraded and the toxin induces cell death. TA genes are ubiquitous in clinical isolates of certain drug-resistant bacteria, and it has been postulated that compounds that disrupt the TA interaction could free the toxin to kill the bac
- High-throughput multiplex microsphere dose response for inhibitors of toxin protease, specifically Botulinum neurotoxin light chain A protease, compounds from Cherry Pick 01 University of New Mexico Assay Overview: Assay Support: 1 R03 MH093184-01A1 Project Title: High-throughput multiplex microsphere screening for toxin protease inhibitors Assay Provider: Steven Graves Ph.D. Screening Center/PI: UNMCMD/ Larry Sklar Ph.D. Lead Biologist: Bruce Edwards Ph.D., Screening Operations Team: Jingshu Zhu, Mark Carter MS, Kristine Gouveia MS, Matthew Garcia Chemistry Center PI: Craig W. Lindsley Chemistry Lead: Kyle Emmitte Specialized Chemistry Center: Vanderbilt Specialized Chemistry Center For Accelerated Probe Development Proteases regulate many biological pathways that include: coagulation, immune system activation, metastasis, and viral life cycles. Within the larger set of proteases, pharmaceutical development for the proteases of the two-part bacterial toxins of Clostridium botulinum and Bacillus anthracis is of great interest due to their role in natural disease and biothreat scenarios (1-4). Botulinum Neurotoxin A Light Chain (BoNTALC) is also
- High-throughput multiplex microsphere dose response for inhibitors of toxin protease, specifically Botulinum neurotoxin light chain A protease, compounds from Cherry Pick 02 University of New Mexico Assay Overview: Assay Support: 1 R03 MH093184-01A1 Project Title: High-throughput multiplex microsphere screening for toxin protease inhibitors Assay Provider: Steven Graves Ph.D. Screening Center/PI: UNMCMD/ Larry Sklar Ph.D. Lead Biologist: Bruce Edwards Ph.D., Screening Operations Team: Jingshu Zhu, Mark Carter MS, Kristine Gouveia MS, Matthew Garcia Chemistry Center PI: Craig W. Lindsley Chemistry Lead: Kyle Emmitte Specialized Chemistry Center: Vanderbilt Specialized Chemistry Center For Accelerated Probe Development Proteases regulate many biological pathways that include: coagulation, immune system activation, metastasis, and viral life cycles. Within the larger set of proteases, pharmaceutical development for the proteases of the two-part bacterial toxins of Clostridium botulinum and Bacillus anthracis is of great interest due to their role in natural disease and biothreat scenarios (1-4). Botulinum Neurotoxin A Light Chain (BoNTALC) is also
- High-throughput multiplex microsphere dose response for inhibitors of toxin protease, specifically Botulinum neurotoxin light chain A protease, compounds from Powder Set 01 University of New Mexico Assay Overview: Assay Support: 1 R03 MH093184-01A1 Project Title: High-throughput multiplex microsphere screening for toxin protease inhibitors Assay Provider: Steven Graves Ph.D. Screening Center/PI: UNMCMD/ Larry Sklar Ph.D. Lead Biologist: Bruce Edwards Ph.D., Screening Operations Team: Jingshu Zhu, Mark Carter MS, Kristine Gouveia MS, Matthew Garcia Chemistry Center PI: Craig W. Lindsley Chemistry Lead: Kyle Emmitte Specialized Chemistry Center: Vanderbilt Specialized Chemistry Center For Accelerated Probe Development Proteases regulate many biological pathways that include: coagulation, immune system activation, metastasis, and viral life cycles. Within the larger set of proteases, pharmaceutical development for the proteases of the two-part bacterial toxins of Clostridium botulinum and Bacillus anthracis is of great interest due to their role in natural disease and biothreat scenarios (1-4). Botulinum Neurotoxin A Light Chain (BoNTALC) is also
- High-throughput multiplex microsphere dose response for inhibitors of toxin protease, specifically Botulinum neurotoxin light chain F protease, compounds from Cherry Pick 01 University of New Mexico Assay Overview: Assay Support: 1 R03 MH093184-01A1 Project Title: High-throughput multiplex microsphere screening for toxin protease inhibitors Assay Provider: Steven Graves Ph.D. Screening Center/PI: UNMCMD/ Larry Sklar Ph.D. Lead Biologist: Bruce Edwards Ph.D., Screening Operations Team: Jingshu Zhu, Mark Carter MS, Kristine Gouveia MS, Matthew Garcia Chemistry Center PI: Craig W. Lindsley Chemistry Lead: Kyle Emmitte Specialized Chemistry Center: Vanderbilt Specialized Chemistry Center For Accelerated Probe Development Proteases regulate many biological pathways that include: coagulation, immune system activation, metastasis, and viral life cycles. Within the larger set of proteases, pharmaceutical development for the proteases of the two-part bacterial toxins of Clostridium botulinum and Bacillus anthracis is of great interest due to their role in natural disease and biothreat scenarios (1-4). Botulinum Neurotoxin A Light Chain (BoNTALC) is also
- High-throughput multiplex microsphere dose response for inhibitors of toxin protease, specifically Botulinum neurotoxin light chain F protease, compounds from Cherry Pick 02 University of New Mexico Assay Overview: Assay Support: 1 R03 MH093184-01A1 Project Title: High-throughput multiplex microsphere screening for toxin protease inhibitors Assay Provider: Steven Graves Ph.D. Screening Center/PI: UNMCMD/ Larry Sklar Ph.D. Lead Biologist: Bruce Edwards Ph.D., Screening Operations Team: Jingshu Zhu, Mark Carter MS, Kristine Gouveia MS, Matthew Garcia Chemistry Center PI: Craig W. Lindsley Chemistry Lead: Kyle Emmitte Specialized Chemistry Center: Vanderbilt Specialized Chemistry Center For Accelerated Probe Development Proteases regulate many biological pathways that include: coagulation, immune system activation, metastasis, and viral life cycles. Within the larger set of proteases, pharmaceutical development for the proteases of the two-part bacterial toxins of Clostridium botulinum and Bacillus anthracis is of great interest due to their role in natural disease and biothreat scenarios (1-4). Botulinum Neurotoxin A Light Chain (BoNTALC) is also
- High-throughput multiplex microsphere dose response for inhibitors of toxin protease, specifically Botulinum neurotoxin light chain F protease, compounds from Powder Set 01 University of New Mexico Assay Overview: Assay Support: 1 R03 MH093184-01A1 Project Title: High-throughput multiplex microsphere screening for toxin protease inhibitors Assay Provider: Steven Graves Ph.D. Screening Center/PI: UNMCMD/ Larry Sklar Ph.D. Lead Biologist: Bruce Edwards Ph.D., Screening Operations Team: Jingshu Zhu, Mark Carter MS, Kristine Gouveia MS, Matthew Garcia Chemistry Center PI: Craig W. Lindsley Chemistry Lead: Kyle Emmitte Specialized Chemistry Center: Vanderbilt Specialized Chemistry Center For Accelerated Probe Development Proteases regulate many biological pathways that include: coagulation, immune system activation, metastasis, and viral life cycles. Within the larger set of proteases, pharmaceutical development for the proteases of the two-part bacterial toxins of Clostridium botulinum and Bacillus anthracis is of great interest due to their role in natural disease and biothreat scenarios (1-4). Botulinum Neurotoxin A Light Chain (BoNTALC) is also
- High-throughput multiplex microsphere dose response for inhibitors of toxin protease, specifically Lethal Factor protease, compounds from Cherry Pick 01 University of New Mexico Assay Overview: Assay Support: 1 R03 MH093184-01A1 Project Title: High-throughput multiplex microsphere screening for toxin protease inhibitors Assay Provider: Steven Graves Ph.D. Screening Center/PI: UNMCMD/ Larry Sklar Ph.D. Lead Biologist: Bruce Edwards Ph.D., Screening Operations Team: Jingshu Zhu, Mark Carter MS, Kristine Gouveia MS, Matthew Garcia Chemistry Center PI: Craig W. Lindsley Chemistry Lead: Kyle Emmitte Specialized Chemistry Center: Vanderbilt Specialized Chemistry Center For Accelerated Probe Development Proteases regulate many biological pathways that include: coagulation, immune system activation, metastasis, and viral life cycles. Within the larger set of proteases, pharmaceutical development for the proteases of the two-part bacterial toxins of Clostridium botulinum and Bacillus anthracis is of great interest due to their role in natural disease and biothreat scenarios (1-4). Botulinum Neurotoxin A Light Chain (BoNTALC) is also
- High-throughput multiplex microsphere dose response for inhibitors of toxin protease, specifically Lethal Factor protease, compounds from Cherry Pick 02 University of New Mexico Assay Overview: Assay Support: 1 R03 MH093184-01A1 Project Title: High-throughput multiplex microsphere screening for toxin protease inhibitors Assay Provider: Steven Graves Ph.D. Screening Center/PI: UNMCMD/ Larry Sklar Ph.D. Lead Biologist: Bruce Edwards Ph.D., Screening Operations Team: Jingshu Zhu, Mark Carter MS, Kristine Gouveia MS, Matthew Garcia Chemistry Center PI: Craig W. Lindsley Chemistry Lead: Kyle Emmitte Specialized Chemistry Center: Vanderbilt Specialized Chemistry Center For Accelerated Probe Development Proteases regulate many biological pathways that include: coagulation, immune system activation, metastasis, and viral life cycles. Within the larger set of proteases, pharmaceutical development for the proteases of the two-part bacterial toxins of Clostridium botulinum and Bacillus anthracis is of great interest due to their role in natural disease and biothreat scenarios (1-4). Botulinum Neurotoxin A Light Chain (BoNTALC) is also
- High-throughput multiplex microsphere dose response for inhibitors of toxin protease, specifically Lethal Factor protease, compounds from Powder Set 01 University of New Mexico Assay Overview: Assay Support: 1 R03 MH093184-01A1 Project Title: High-throughput multiplex microsphere screening for toxin protease inhibitors Assay Provider: Steven Graves Ph.D. Screening Center/PI: UNMCMD/ Larry Sklar Ph.D. Lead Biologist: Bruce Edwards Ph.D., Screening Operations Team: Jingshu Zhu, Mark Carter MS, Kristine Gouveia MS, Matthew Garcia Chemistry Center PI: Craig W. Lindsley Chemistry Lead: Kyle Emmitte Specialized Chemistry Center: Vanderbilt Specialized Chemistry Center For Accelerated Probe Development Proteases regulate many biological pathways that include: coagulation, immune system activation, metastasis, and viral life cycles. Within the larger set of proteases, pharmaceutical development for the proteases of the two-part bacterial toxins of Clostridium botulinum and Bacillus anthracis is of great interest due to their role in natural disease and biothreat scenarios (1-4). Botulinum Neurotoxin A Light Chain (BoNTALC) is also
- Dose Response of Small Molecules that Regulate V-ATPase Proton Transport in Yeast using pHLuorin, Cherry Pick 1 University of New Mexico Assay Overview: Assay Support: 1 R03 DA031666-01A1 Project Title: Flow Cytometry HTS of Small Molecules that Regulate V-ATPase Proton Transport in Yeast Assay Provider: Karlett Parra Ph.D Lead Biologist: Mark Carter MS Chemistry Center/ PI: Specialized Chemistry Center: Assay Implementation: Travis Houston, Keon Ahghar, Stephanie Chavez, Dominique Perez, Matthew Garcia, Sahba Charkhzarrin, Anna Waller Ph.D, Annette Evangelisti Ph.D Assay Background and Significance: Distributed among the endomembrane system of all eukaryotic cells, V-ATPase proton pumps are responsible for acidification of intracellular compartments. V-ATPases maintain the low pH necessary for endocytic and exocytic vesicular transport, zymogen activation, and protein sorting and degradation. Enveloped viruses, such as influenza virus, as well as toxins, such as diphtheria toxin, enter cells via acidic endosomal compartments, in which low pH is maintained by V-ATPases. Because the pH influe
- Dose Response of Small Molecules that Regulate V-ATPase Proton Transport in Yeast using pHLuorin, Cherry Pick 2 University of New Mexico Assay Overview: Assay Support: 1 R03 DA031666-01A1 Project Title: Flow Cytometry HTS of Small Molecules that Regulate V-ATPase Proton Transport in Yeast Assay Provider: Karlett Parra Ph.D Lead Biologist: Mark Carter MS Chemistry Center/ PI: Specialized Chemistry Center: Assay Implementation: Chun-Yuan Chan, Stephanie Chavez, Dominique Perez, Matthew Garcia, Terry Foutz, Anna Waller Ph.D, Annette Evangelisti Ph.D, Gergely Zahoransky-Kohalmi Assay Background and Significance: Distributed among the endomembrane system of all eukaryotic cells, V-ATPase proton pumps are responsible for acidification of intracellular compartments. V-ATPases maintain the low pH necessary for endocytic and exocytic vesicular transport, zymogen activation, and protein sorting and degradation. Enveloped viruses, such as influenza virus, as well as toxins, such as diphtheria toxin, enter cells via acidic endosomal compartments, in which low pH is maintained by V-ATPases. Because the
- Dose Response of Small Molecules that Regulate V-ATPase Proton Transport in Yeast using pHLuorin, Powder Set1 University of New Mexico Assay Overview: Assay Support: 1 R03 DA031666-01A1 Project Title: Flow Cytometry HTS of Small Molecules that Regulate V-ATPase Proton Transport in Yeast Assay Provider: Karlett Parra Ph.D Lead Biologist: Mark Carter MS Chemistry Center/ PI: Specialized Chemistry Center: Assay Implementation: Mark Carter, Chun-Yuan Chan, Stephanie Chavez, Dominique Perez, Matthew Garcia, Terry Foutz, Anna Waller Ph.D, Annette Evangelisti Ph.D, Gergely Zahoransky-Kohalmi Assay Background and Significance: Distributed among the endomembrane system of all eukaryotic cells, V-ATPase proton pumps are responsible for acidification of intracellular compartments. V-ATPases maintain the low pH necessary for endocytic and exocytic vesicular transport, zymogen activation, and protein sorting and degradation. Enveloped viruses, such as influenza virus, as well as toxins, such as diphtheria toxin, enter cells via acidic endosomal compartments, in which low pH is maintained by V-ATPases.
- Dose Response with BCECF Assay for V-ATPase inhibitors that increase vacuolar pH, Powder Set 1 University of New Mexico Assay Overview: Assay Support: 1 R03 DA031666-01A1 Project Title: Flow Cytometry HTS of Small Molecules that Regulate V-ATPase Proton Transport in Yeast Assay Provider: Karlett Parra Ph.D Lead Biologist: Mark Carter MS Chemistry Center/ PI: Specialized Chemistry Center: Assay Implementation: Mark Carter, Chun-Yuan Chan, Matthew Garcia, Terry Foutz, Anna Waller Ph.D, Annette Evangelisti Ph.D, Gergely Zahoransky-Kohalmi Assay Background and Significance: Distributed among the endomembrane system of all eukaryotic cells, V-ATPase proton pumps are responsible for acidification of intracellular compartments. V-ATPases maintain the low pH necessary for endocytic and exocytic vesicular transport, zymogen activation, and protein sorting and degradation. Enveloped viruses, such as influenza virus, as well as toxins, such as diphtheria toxin, enter cells via acidic endosomal compartments, in which low pH is maintained by V-ATPases. Because the pH influences most aspe
- Inhibition Assay Various concentrations of the substances to be tested are dissolved in dimethyl sulphoxide and mixed with an aqueous refludan solution (10 μg/ml). In clear 96-well plates having a flat bottom, 30 μl of citrate plasma (Octapharma) are mixed with 10 μl of the substance dilution. Then, either 20 μl of a solution of a rattlesnake toxin (Russel viper venom (RVV); RVV reagent: Pentapharm 121-06, final concentration 0.6 mU) in an aqueous calcium chloride solution buffer (final concentration of calcium chloride 0.05 M) or 20 μl of the aqueous calcium chloride solution (final concentration of calcium chloride 0.05 M) without RVV reagent (as reference for an unstimulated sample) are added. After addition of 20 μl of ChromozymX substrate (final concentration 1.6 mmol/l, Bachem L-1565, diluted in water) the samples are measured in a SpectraFluor Reader using a measurement filter of 405 nm each minute over a period of 20 minutes.
- MATE2-K IC50 Assay MATE2-K (multidrug and toxin extrusion protein 2) is expressed in the apical membrane in the kidney and mediates the elimination of compounds to urine. MDCK-II cells were maintained in DMEM with low glucose and 10% FBS. Cells passages up to 40 were seeded at 60K±10K cells/well on 96-well, transwell membrane plates approximately 24 hours before transfection. Transport assays were carried out approximately 48 hours after transfection. On assay day, the DMEM was removed, and cells were washed with HBSS. After washing, the cells in each well were pre-incubated with HBSS containing 30 mM NH4Cl and either vehicle, the compound being tested at 6 concentrations ranging from 0.127 μM to 40 AM, or 100 μM cimetidine as a reference inhibitor. The assay plate was then placed in a 37° C. incubator with orbital shaking at approximately 60 RPM for the pre-incubation time of 15 minutes. The pre-incubation solutions were then removed, and cells washed with HBSS once. 100 μL of incubation buffer was added to each well containing HBSS with 10 μM 14[C]-metformin as the probe substrate and either vehicle control, the compound being tested (at 6 concentrations ranging from 0.127 μM to 40 μM) or reference inhibitors. The assay plate was incubated at 37° C. with orbital shaking at approximately 60 RPM for the incubation time of 5 minutes. At end of the 5-minute incubation, 15 μL of dosing solution was removed from each well containing the compound being tested and measured using LC/MS/MS for dose recovery assessment. The assay wells were then washed four times with ice cold PBS. 60 μL cell extraction solution was added to each well and the plate was incubated at 37° C. with orbital shaking at approximately 60 RPM for the 15 minutes. After this incubation, 30 μL was removed from each well, added to 200 μL scintillation fluid, and counted on a 1450 Microbeta (Perkin-Elmer) to measure the probe substrate uptake. Inhibition potential of the compound being tested was calculated by dividing the transporter-mediated uptake rate in presence of the compound being tested or the reference inhibitor by the transporter-mediated uptake rate in presence of vehicle control and fitted to a sigmoidal function to determine the IC50 values.
 US11078214, Compound PTX BDBM511066
US11078214, Compound PTX BDBM511066 HC toxin BDBM198122
HC toxin BDBM198122 BDBM50161873 CHEMBL437624 E[c(RGDyK)]2-PTX conjugate
BDBM50161873 CHEMBL437624 E[c(RGDyK)]2-PTX conjugate SMR001566822 MLS002703014 T-2 TOXIN cid_529495 BDBM93500
SMR001566822 MLS002703014 T-2 TOXIN cid_529495 BDBM93500 BDBM18224 6-[(2,5-dimethoxyphenyl)methyl]-5-methylpyrido[2,3-d]pyrimidine-2,4-diamine Piritrexim PTX [2-amino-6-(2,5-dimethoxybenzyl)-5-methyl-pyrido[2,3-d]pyrimidin-4-yl]amine;2-hydroxyethanesulfonic acid CHEMBL7492
BDBM18224 6-[(2,5-dimethoxyphenyl)methyl]-5-methylpyrido[2,3-d]pyrimidine-2,4-diamine Piritrexim PTX [2-amino-6-(2,5-dimethoxybenzyl)-5-methyl-pyrido[2,3-d]pyrimidin-4-yl]amine;2-hydroxyethanesulfonic acid CHEMBL7492 Joro spider toxin 3 CHEMBL313747 N*1*-(5-{3-[4-(3-Amino-propylamino)-butylamino]-propionylamino}-pentyl)-2-[2-(2,4-dihydroxy-phenyl)-acetylamino]-succinamide 2-Amino-N-{3-[4-(3-amino-propylamino)-butylamino]-propyl}-acetamide BDBM50105843
Joro spider toxin 3 CHEMBL313747 N*1*-(5-{3-[4-(3-Amino-propylamino)-butylamino]-propionylamino}-pentyl)-2-[2-(2,4-dihydroxy-phenyl)-acetylamino]-succinamide 2-Amino-N-{3-[4-(3-amino-propylamino)-butylamino]-propyl}-acetamide BDBM50105843