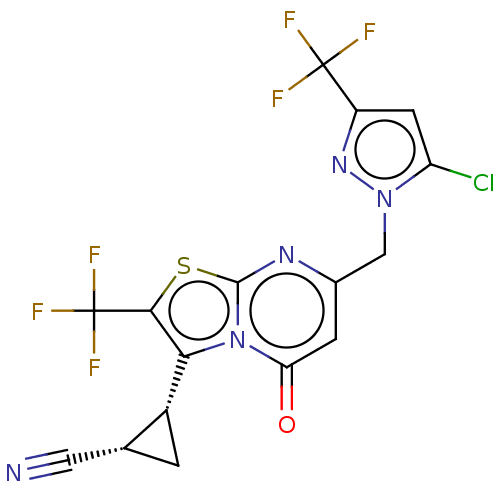
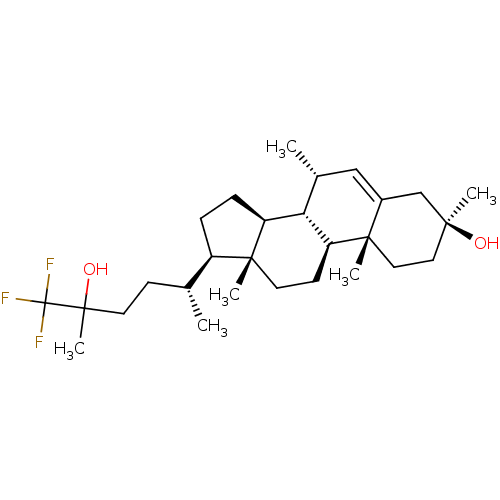
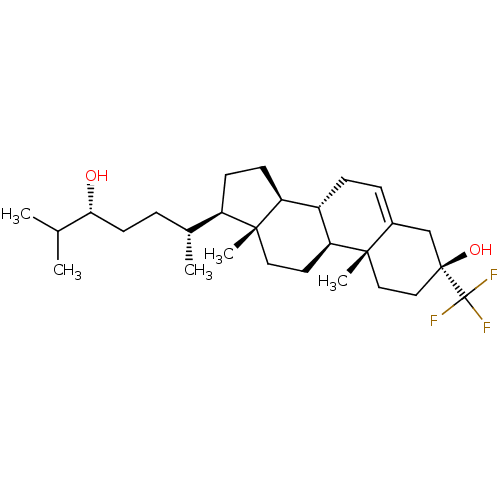
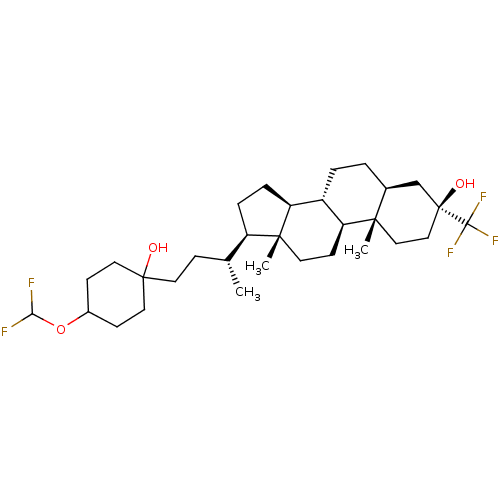
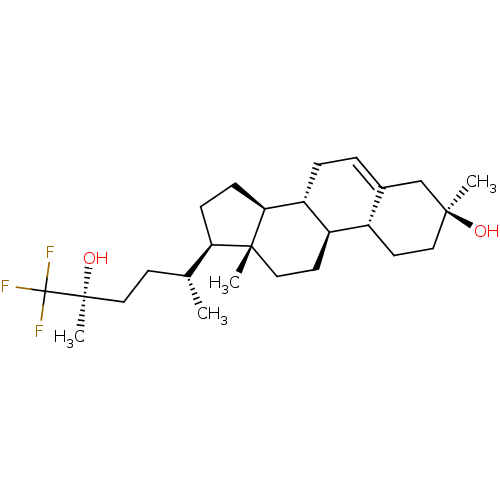
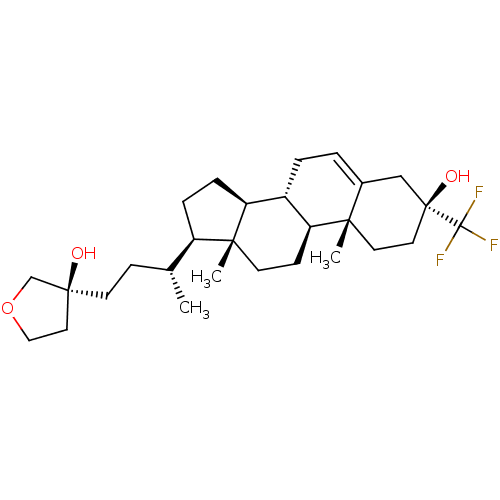
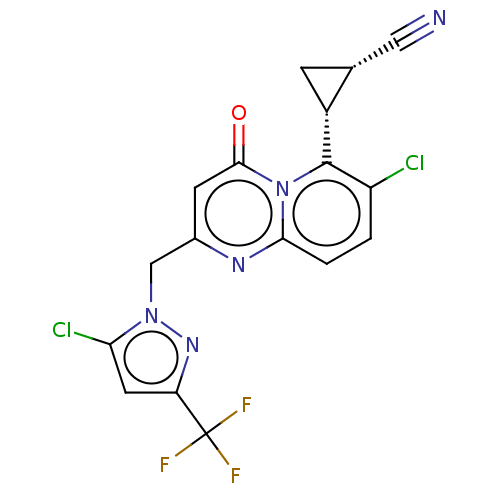
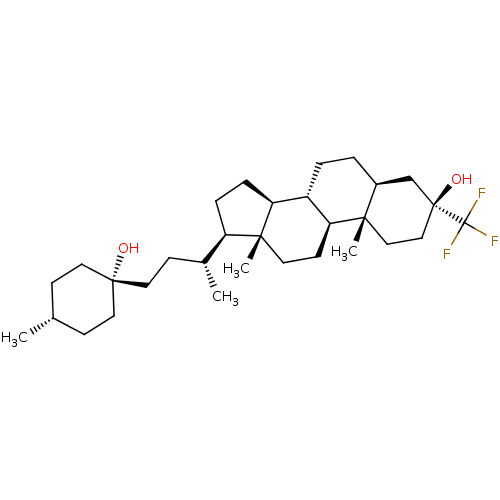
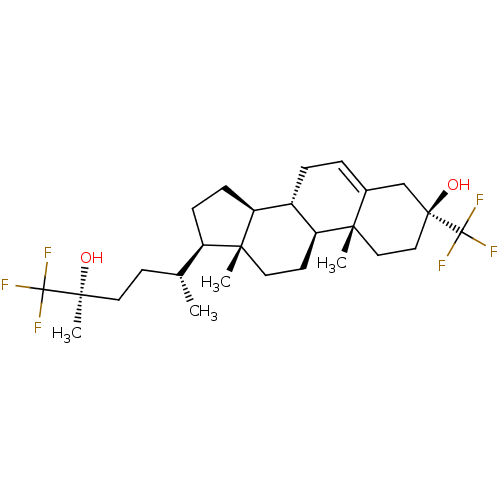
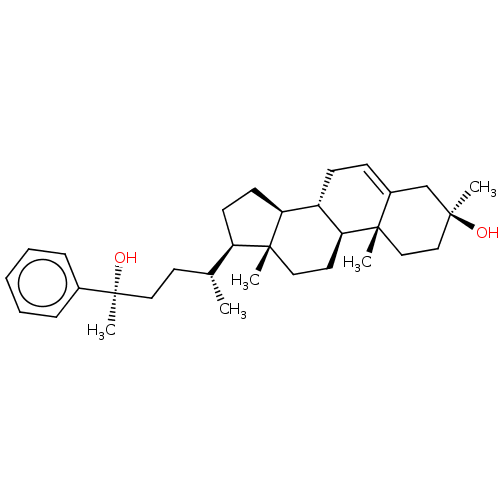
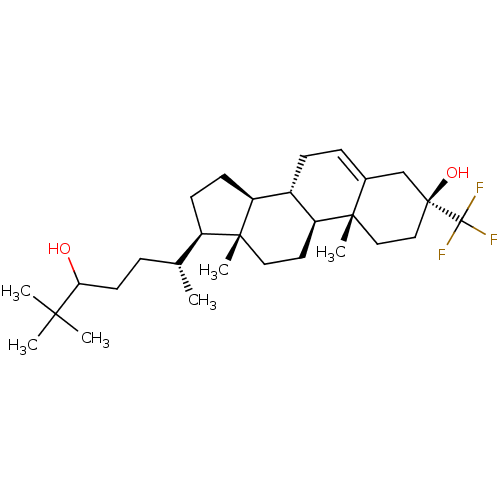
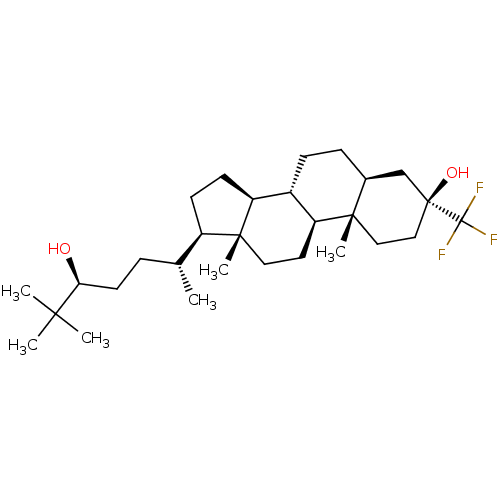
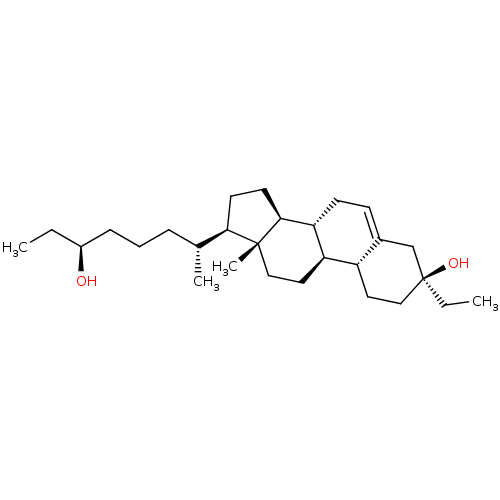
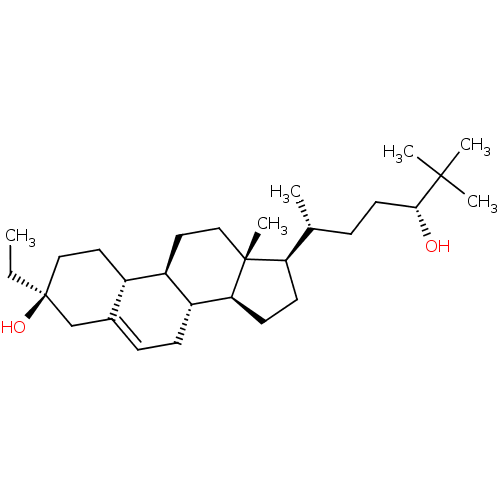
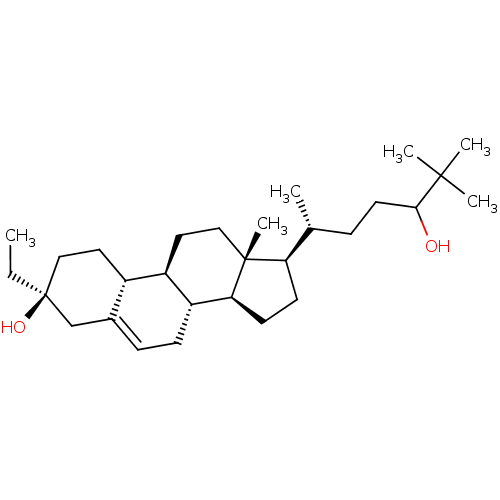
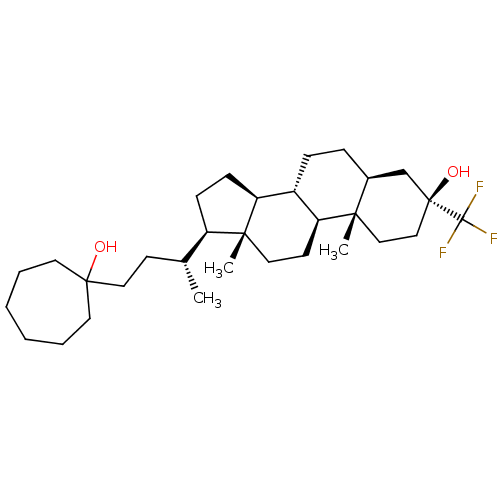
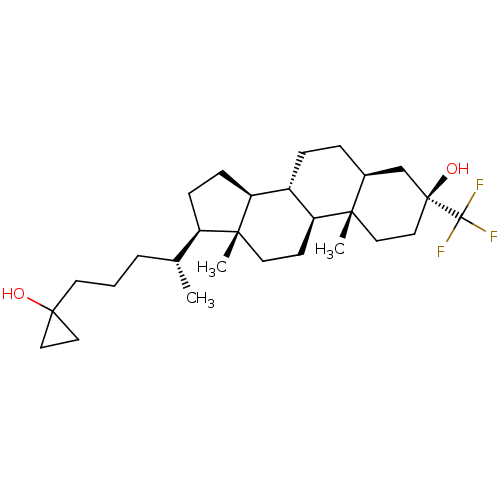
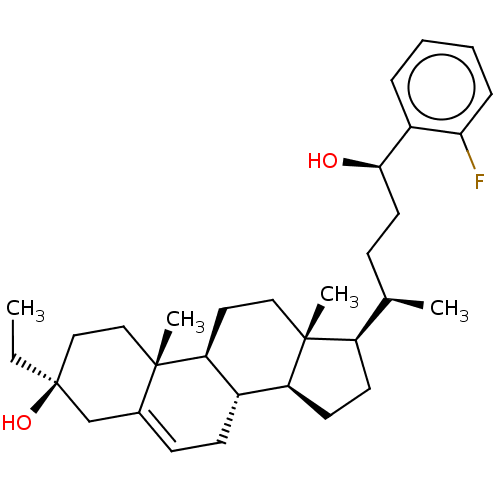
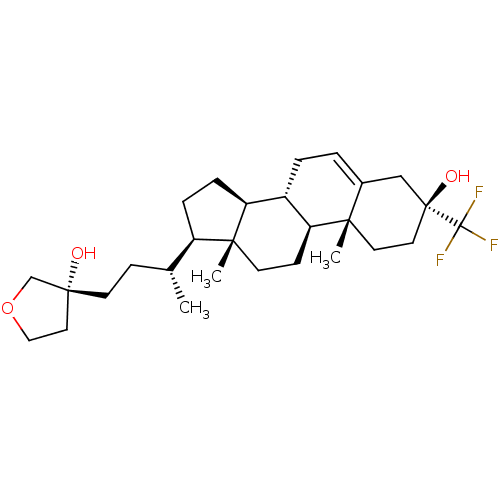
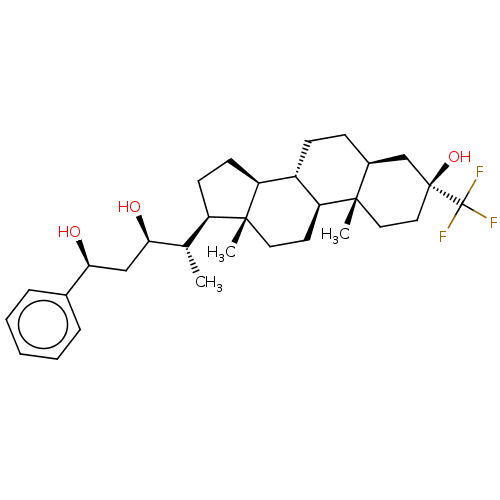
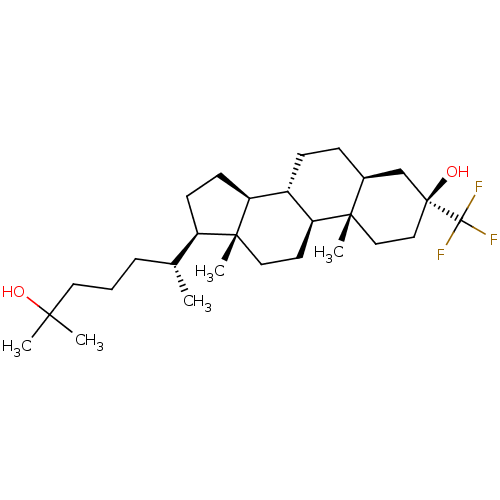
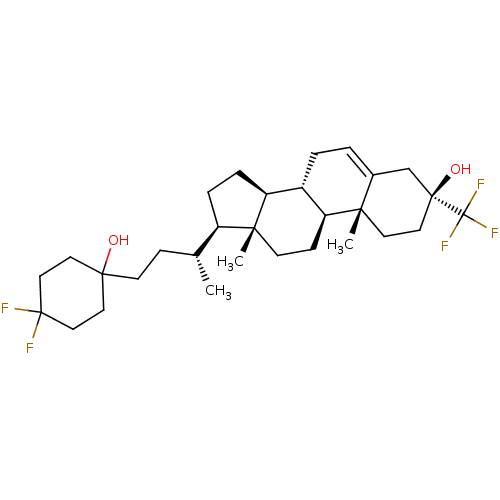
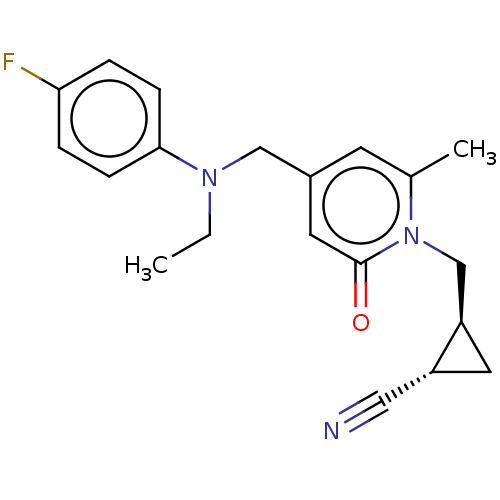
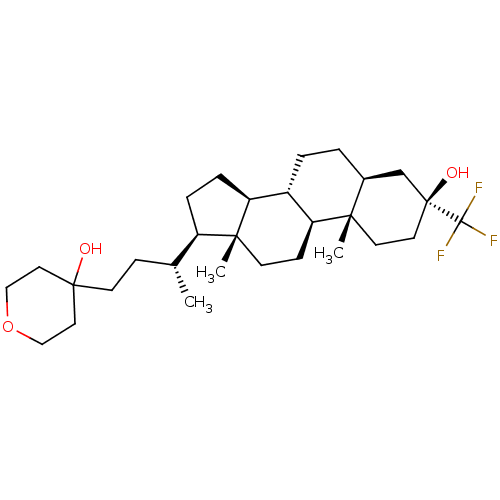
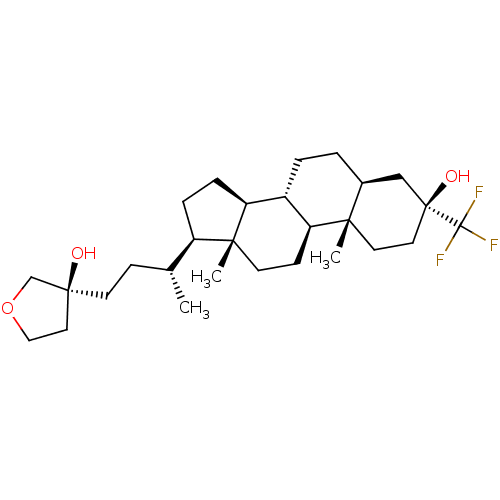
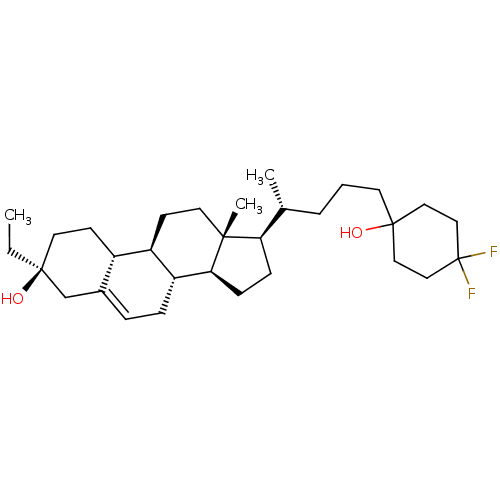
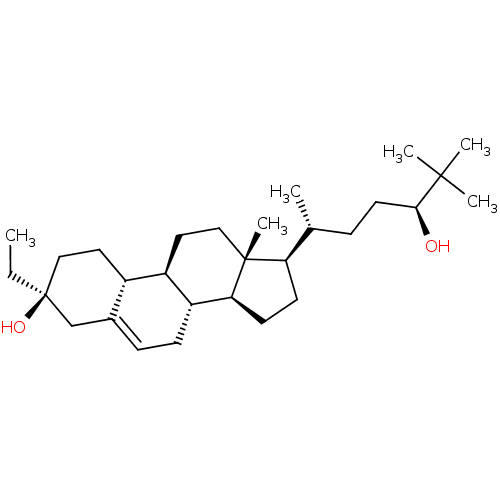
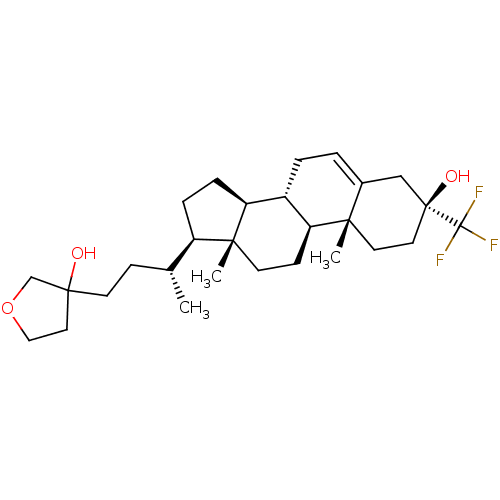
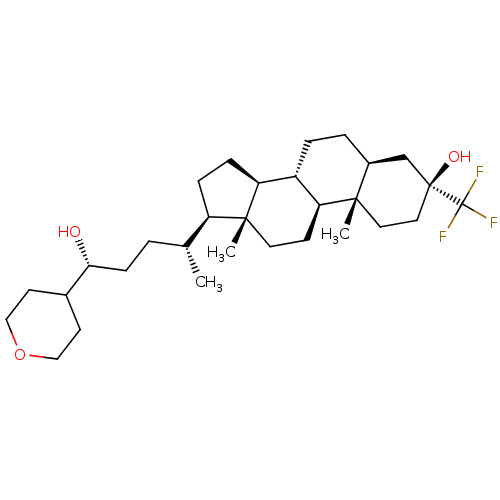
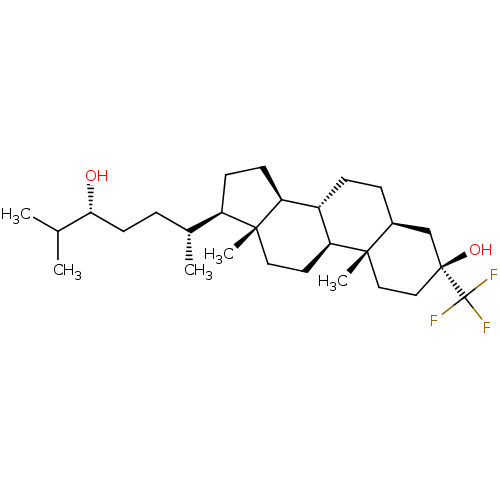
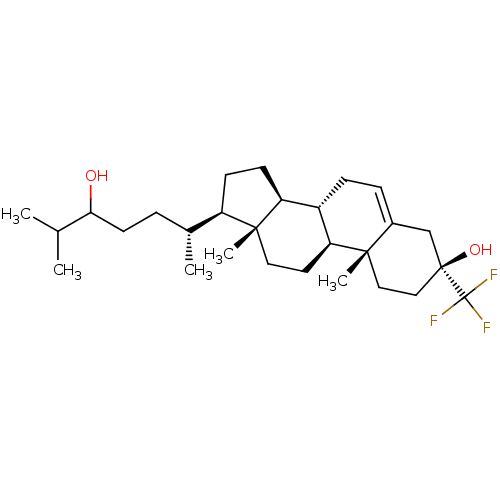
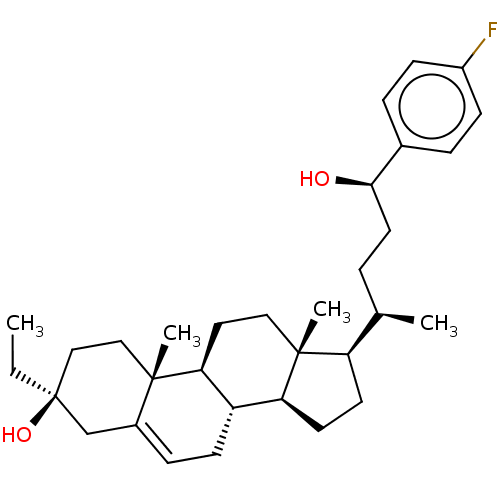
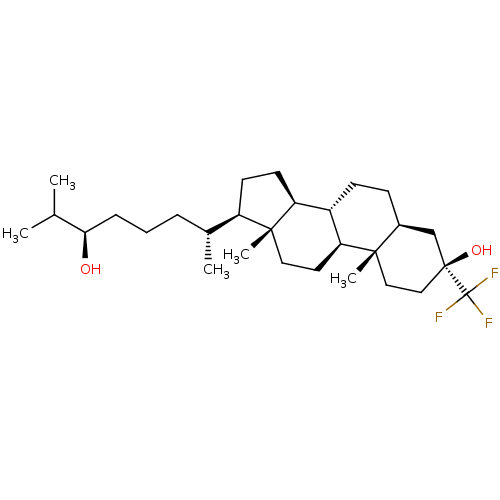
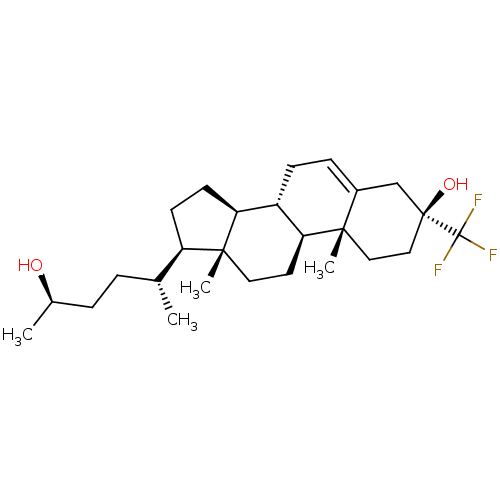
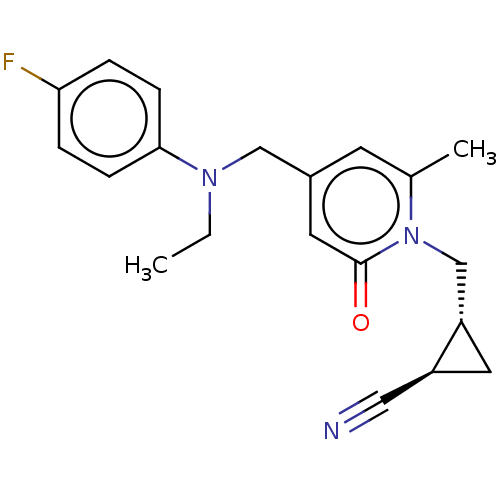
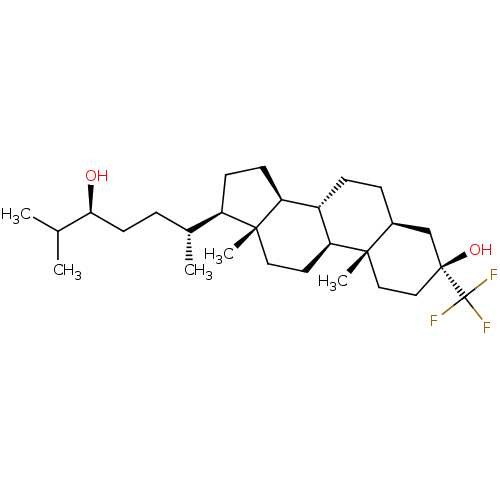
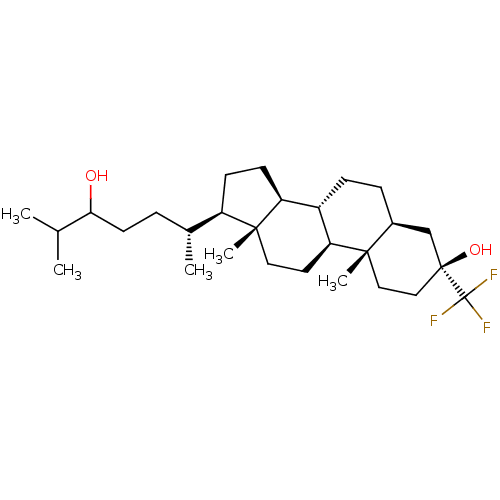
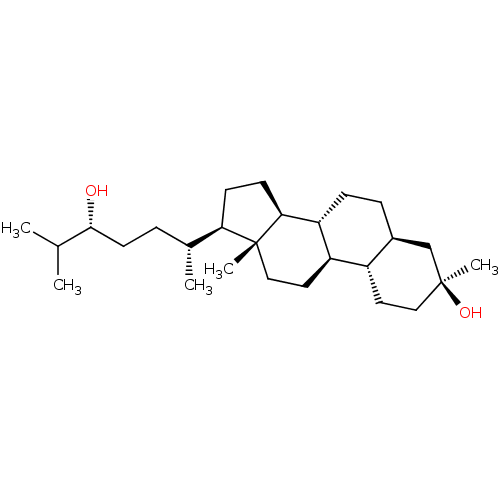
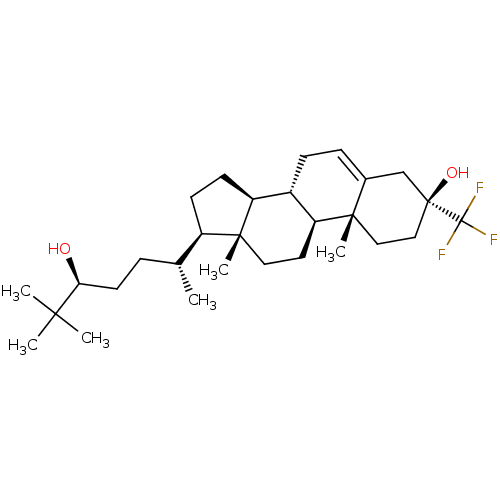
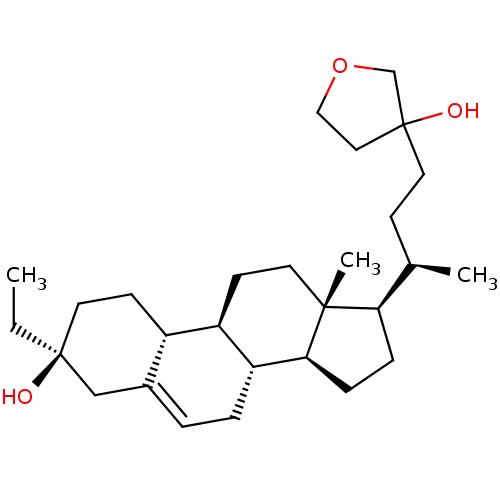
TargetGlutamate receptor ionotropic, NMDA 2A(Homo sapiens (Human))
Takeda Pharmaceutical
Curated by ChEMBL
Takeda Pharmaceutical
Curated by ChEMBL
Affinity DataEC50: 11nMAssay Description:Positive allosteric modulator activity at GluN2A receptor (unknown origin) by FLIPR assayMore data for this Ligand-Target Pair
TargetGlutamate receptor ionotropic, NMDA 2A(Homo sapiens (Human))
Takeda Pharmaceutical
Curated by ChEMBL
Takeda Pharmaceutical
Curated by ChEMBL
Affinity DataEC50: 13nMAssay Description:Cells are transferred as suspension in serum-free medium to the QPatch HTX system and kept in the cell storage tank/stirrer during experiments. All s...More data for this Ligand-Target Pair
TargetGlutamate receptor ionotropic, NMDA 2A(Homo sapiens (Human))
Takeda Pharmaceutical
Curated by ChEMBL
Takeda Pharmaceutical
Curated by ChEMBL
Affinity DataEC50: 31.6nMAssay Description:The whole-cell patch-clamp technique was used to investigate the effects of positive allosteric modulating activity of test compounds on GlunN1/GluN2...More data for this Ligand-Target Pair
TargetGlutamate receptor ionotropic, NMDA 2A(Homo sapiens (Human))
Takeda Pharmaceutical
Curated by ChEMBL
Takeda Pharmaceutical
Curated by ChEMBL
Affinity DataEC50: 33.6nMAssay Description:The whole-cell patch-clamp technique was used to investigate the effects of positive allosteric modulating activity of test compounds on GlunN1/GluN2...More data for this Ligand-Target Pair
TargetGlutamate receptor ionotropic, NMDA 2A(Homo sapiens (Human))
Takeda Pharmaceutical
Curated by ChEMBL
Takeda Pharmaceutical
Curated by ChEMBL
Affinity DataEC50: 34.5nMAssay Description:The whole-cell patch-clamp technique was used to investigate the effects of positive allosteric modulating activity of test compounds on GlunN1/GluN2...More data for this Ligand-Target Pair
TargetGlutamate receptor ionotropic, NMDA 2A(Homo sapiens (Human))
Takeda Pharmaceutical
Curated by ChEMBL
Takeda Pharmaceutical
Curated by ChEMBL
Affinity DataEC50: 35.1nMAssay Description:The whole-cell patch-clamp technique was used to investigate the effects of positive allosteric modulating activity of test compounds on GlunN1/GluN2...More data for this Ligand-Target Pair
TargetGlutamate receptor ionotropic, NMDA 2A(Homo sapiens (Human))
Takeda Pharmaceutical
Curated by ChEMBL
Takeda Pharmaceutical
Curated by ChEMBL
Affinity DataEC50: 39nMAssay Description:Positive allosteric modulator activity at GluN2A receptor (unknown origin) by FLIPR assayMore data for this Ligand-Target Pair
TargetGlutamate receptor ionotropic, NMDA 2A(Homo sapiens (Human))
Takeda Pharmaceutical
Curated by ChEMBL
Takeda Pharmaceutical
Curated by ChEMBL
Affinity DataEC50: 53.4nMAssay Description:The whole-cell patch-clamp technique was used to investigate the effects of positive allosteric modulating activity of test compounds on GlunN1/GluN2...More data for this Ligand-Target Pair
TargetGlutamate receptor ionotropic, NMDA 2A(Homo sapiens (Human))
Takeda Pharmaceutical
Curated by ChEMBL
Takeda Pharmaceutical
Curated by ChEMBL
Affinity DataEC50: 61nMAssay Description:The whole-cell patch-clamp technique was used to investigate the effects of positive allosteric modulating activity of test compounds on GlunN1/GluN2...More data for this Ligand-Target Pair
TargetGlutamate receptor ionotropic, NMDA 2A(Homo sapiens (Human))
Takeda Pharmaceutical
Curated by ChEMBL
Takeda Pharmaceutical
Curated by ChEMBL
Affinity DataEC50: 61.4nMAssay Description:The whole-cell patch-clamp technique was used to investigate the effects of positive allosteric modulating activity of test compounds on GlunN1/GluN2...More data for this Ligand-Target Pair
TargetGlutamate receptor ionotropic, NMDA 2A(Homo sapiens (Human))
Takeda Pharmaceutical
Curated by ChEMBL
Takeda Pharmaceutical
Curated by ChEMBL
Affinity DataEC50: 63.3nMAssay Description:The whole-cell patch-clamp technique was used to investigate the effects of positive allosteric modulating activity of test compounds on GlunN1/GluN2...More data for this Ligand-Target Pair
TargetGlutamate receptor ionotropic, NMDA 2A(Homo sapiens (Human))
Takeda Pharmaceutical
Curated by ChEMBL
Takeda Pharmaceutical
Curated by ChEMBL
Affinity DataEC50: 65.8nMAssay Description:The whole-cell patch-clamp technique was used to investigate the effects of positive allosteric modulating activity of test compounds on GlunN1/GluN2...More data for this Ligand-Target Pair
TargetGlutamate receptor ionotropic, NMDA 2A(Homo sapiens (Human))
Takeda Pharmaceutical
Curated by ChEMBL
Takeda Pharmaceutical
Curated by ChEMBL
Affinity DataEC50: 66.4nMAssay Description:The whole-cell patch-clamp technique was used to investigate the effects of positive allosteric modulating activity of test compounds on GlunN1/GluN2...More data for this Ligand-Target Pair
TargetGlutamate receptor ionotropic, NMDA 2A(Homo sapiens (Human))
Takeda Pharmaceutical
Curated by ChEMBL
Takeda Pharmaceutical
Curated by ChEMBL
Affinity DataEC50: 67.6nMAssay Description:The whole-cell patch-clamp technique was used to investigate the effects of positive allosteric modulating activity of test compounds on GlunN1/GluN2...More data for this Ligand-Target Pair
TargetGlutamate receptor ionotropic, NMDA 2A(Homo sapiens (Human))
Takeda Pharmaceutical
Curated by ChEMBL
Takeda Pharmaceutical
Curated by ChEMBL
Affinity DataEC50: 69.7nMAssay Description:The whole-cell patch-clamp technique was used to investigate the effects of positive allosteric modulating activity of test compounds on GlunN1/GluN2...More data for this Ligand-Target Pair
TargetGlutamate receptor ionotropic, NMDA 2A(Homo sapiens (Human))
Takeda Pharmaceutical
Curated by ChEMBL
Takeda Pharmaceutical
Curated by ChEMBL
Affinity DataEC50: 73.4nMAssay Description:The whole-cell patch-clamp technique was used to investigate the effects of positive allosteric modulating activity of test compounds on GlunN1/GluN2...More data for this Ligand-Target Pair
TargetGlutamate receptor ionotropic, NMDA 2A(Homo sapiens (Human))
Takeda Pharmaceutical
Curated by ChEMBL
Takeda Pharmaceutical
Curated by ChEMBL
Affinity DataEC50: 87nMAssay Description:The whole-cell patch-clamp technique was used to investigate the effects of positive allosteric modulating activity of test compounds on GlunN1/GluN2...More data for this Ligand-Target Pair
TargetGlutamate receptor ionotropic, NMDA 2A(Homo sapiens (Human))
Takeda Pharmaceutical
Curated by ChEMBL
Takeda Pharmaceutical
Curated by ChEMBL
Affinity DataEC50: 88.1nMAssay Description:The whole-cell patch-clamp technique was used to investigate the effects of positive allosteric modulating activity of test compounds on GlunN1/GluN2...More data for this Ligand-Target Pair
TargetGlutamate receptor ionotropic, NMDA 2A(Homo sapiens (Human))
Takeda Pharmaceutical
Curated by ChEMBL
Takeda Pharmaceutical
Curated by ChEMBL
Affinity DataEC50: 91.9nMAssay Description:The whole-cell patch-clamp technique was used to investigate the effects of positive allosteric modulating activity of test compounds on GlunN1/GluN2...More data for this Ligand-Target Pair
TargetGlutamate receptor ionotropic, NMDA 2A(Homo sapiens (Human))
Takeda Pharmaceutical
Curated by ChEMBL
Takeda Pharmaceutical
Curated by ChEMBL
Affinity DataEC50: 95.9nMAssay Description:The whole-cell patch-clamp technique was used to investigate the effects of positive allosteric modulating activity of test compounds on GlunN1/GluN2...More data for this Ligand-Target Pair
TargetGlutamate receptor ionotropic, NMDA 2A(Homo sapiens (Human))
Takeda Pharmaceutical
Curated by ChEMBL
Takeda Pharmaceutical
Curated by ChEMBL
Affinity DataEC50: 96.7nMAssay Description:The whole-cell patch-clamp technique was used to investigate the effects of positive allosteric modulating activity of test compounds on GlunN1/GluN2...More data for this Ligand-Target Pair
TargetGlutamate receptor ionotropic, NMDA 2A(Homo sapiens (Human))
Takeda Pharmaceutical
Curated by ChEMBL
Takeda Pharmaceutical
Curated by ChEMBL
Affinity DataEC50: <100nMAssay Description:Whole-Cell Patch Clamp of Mammalian Cells (Ionworks Barracuda (IWB)):The whole-cell patch-clamp technique was used to investigate the effects of posi...More data for this Ligand-Target Pair
TargetGlutamate receptor ionotropic, NMDA 2A(Homo sapiens (Human))
Takeda Pharmaceutical
Curated by ChEMBL
Takeda Pharmaceutical
Curated by ChEMBL
Affinity DataEC50: <100nMAssay Description:Whole-Cell Patch Clamp of Mammalian Cells (Ionworks Barracuda (IWB)):The whole-cell patch-clamp technique was used to investigate the effects of posi...More data for this Ligand-Target Pair
TargetGlutamate receptor ionotropic, NMDA 2A(Homo sapiens (Human))
Takeda Pharmaceutical
Curated by ChEMBL
Takeda Pharmaceutical
Curated by ChEMBL
Affinity DataEC50: <100nMAssay Description:Whole-Cell Patch Clamp of Mammalian Cells (Ionworks Barracuda (IWB)):The whole-cell patch-clamp technique was used to investigate the effects of posi...More data for this Ligand-Target Pair
TargetGlutamate receptor ionotropic, NMDA 2A(Homo sapiens (Human))
Takeda Pharmaceutical
Curated by ChEMBL
Takeda Pharmaceutical
Curated by ChEMBL
Affinity DataEC50: 102nMAssay Description:The whole-cell patch-clamp technique was used to investigate the effects of positive allosteric modulating activity of test compounds on GlunN1/GluN2...More data for this Ligand-Target Pair
TargetGlutamate receptor ionotropic, NMDA 2A(Homo sapiens (Human))
Takeda Pharmaceutical
Curated by ChEMBL
Takeda Pharmaceutical
Curated by ChEMBL
Affinity DataEC50: 102nMAssay Description:The whole-cell patch-clamp technique was used to investigate the effects of positive allosteric modulating activity of test compounds on GlunN1/GluN2...More data for this Ligand-Target Pair
TargetGlutamate receptor ionotropic, NMDA 2A(Homo sapiens (Human))
Takeda Pharmaceutical
Curated by ChEMBL
Takeda Pharmaceutical
Curated by ChEMBL
Affinity DataEC50: 104nMAssay Description:The whole-cell patch-clamp technique was used to investigate the effects of positive allosteric modulating activity of test compounds on GlunN1/GluN2...More data for this Ligand-Target Pair
TargetGlutamate receptor ionotropic, NMDA 2A(Homo sapiens (Human))
Takeda Pharmaceutical
Curated by ChEMBL
Takeda Pharmaceutical
Curated by ChEMBL
Affinity DataEC50: 109nMAssay Description:The whole-cell patch-clamp technique was used to investigate the effects of positive allosteric modulating activity of test compounds on GlunN1/GluN2...More data for this Ligand-Target Pair
TargetGlutamate receptor ionotropic, NMDA 2A(Homo sapiens (Human))
Takeda Pharmaceutical
Curated by ChEMBL
Takeda Pharmaceutical
Curated by ChEMBL
Affinity DataEC50: 110nMAssay Description:Positive allosteric modulator activity at GluN2A NMDA receptor (unknown origin) expressed in CHO cells co-expressing GluN1a in the presence of L-glut...More data for this Ligand-Target Pair
Ligand Info
TargetGlutamate receptor ionotropic, NMDA 2A(Homo sapiens (Human))
Takeda Pharmaceutical
Curated by ChEMBL
Takeda Pharmaceutical
Curated by ChEMBL
Affinity DataEC50: 117nMAssay Description:The whole-cell patch-clamp technique was used to investigate the effects of positive allosteric modulating activity of test compounds on GlunN1/GluN2...More data for this Ligand-Target Pair
TargetGlutamate receptor ionotropic, NMDA 2A(Homo sapiens (Human))
Takeda Pharmaceutical
Curated by ChEMBL
Takeda Pharmaceutical
Curated by ChEMBL
Affinity DataEC50: 119nMAssay Description:The whole-cell patch-clamp technique was used to investigate the effects of positive allosteric modulating activity of test compounds on GlunN1/GluN2...More data for this Ligand-Target Pair
TargetGlutamate receptor ionotropic, NMDA 2A(Homo sapiens (Human))
Takeda Pharmaceutical
Curated by ChEMBL
Takeda Pharmaceutical
Curated by ChEMBL
Affinity DataEC50: 120nMAssay Description:The whole-cell patch-clamp technique was used to investigate the effects of positive allosteric modulating activity of test compounds on GlunN1/GluN2...More data for this Ligand-Target Pair
TargetGlutamate receptor ionotropic, NMDA 2A(Homo sapiens (Human))
Takeda Pharmaceutical
Curated by ChEMBL
Takeda Pharmaceutical
Curated by ChEMBL
Affinity DataEC50: 124nMAssay Description:The whole-cell patch-clamp technique was used to investigate the effects of positive allosteric modulating activity of test compounds on GlunN1/GluN2...More data for this Ligand-Target Pair
TargetGlutamate receptor ionotropic, NMDA 2A(Homo sapiens (Human))
Takeda Pharmaceutical
Curated by ChEMBL
Takeda Pharmaceutical
Curated by ChEMBL
Affinity DataEC50: 124nMAssay Description:The whole-cell patch-clamp technique was used to investigate the effects of positive allosteric modulating activity of test compounds on GlunN1/GluN2...More data for this Ligand-Target Pair
TargetGlutamate receptor ionotropic, NMDA 2A(Homo sapiens (Human))
Takeda Pharmaceutical
Curated by ChEMBL
Takeda Pharmaceutical
Curated by ChEMBL
Affinity DataEC50: 125nMAssay Description:The whole-cell patch-clamp technique was used to investigate the effects of positive allosteric modulating activity of test compounds on GlunN1/GluN2...More data for this Ligand-Target Pair
TargetGlutamate receptor ionotropic, NMDA 2A(Homo sapiens (Human))
Takeda Pharmaceutical
Curated by ChEMBL
Takeda Pharmaceutical
Curated by ChEMBL
Affinity DataEC50: 129nMAssay Description:The whole-cell patch-clamp technique was used to investigate the effects of positive allosteric modulating activity of test compounds on GlunN1/GluN2...More data for this Ligand-Target Pair
TargetGlutamate receptor ionotropic, NMDA 2A(Homo sapiens (Human))
Takeda Pharmaceutical
Curated by ChEMBL
Takeda Pharmaceutical
Curated by ChEMBL
Affinity DataEC50: 131nMAssay Description:The whole-cell patch-clamp technique was used to investigate the effects of positive allosteric modulating activity of test compounds on GlunN1/GluN2...More data for this Ligand-Target Pair
TargetGlutamate receptor ionotropic, NMDA 2A(Homo sapiens (Human))
Takeda Pharmaceutical
Curated by ChEMBL
Takeda Pharmaceutical
Curated by ChEMBL
Affinity DataEC50: 135nMAssay Description:The whole-cell patch-clamp technique was used to investigate the effects of positive allosteric modulating activity of test compounds on GlunN1/GluN2...More data for this Ligand-Target Pair
TargetGlutamate receptor ionotropic, NMDA 2A(Homo sapiens (Human))
Takeda Pharmaceutical
Curated by ChEMBL
Takeda Pharmaceutical
Curated by ChEMBL
Affinity DataEC50: 145nMAssay Description:The whole-cell patch-clamp technique was used to investigate the effects of positive allosteric modulating activity of test compounds on GlunN1/GluN2...More data for this Ligand-Target Pair
TargetGlutamate receptor ionotropic, NMDA 2A(Homo sapiens (Human))
Takeda Pharmaceutical
Curated by ChEMBL
Takeda Pharmaceutical
Curated by ChEMBL
Affinity DataEC50: 147nMAssay Description:The whole-cell patch-clamp technique was used to investigate the effects of positive allosteric modulating activity of test compounds on GlunN1/GluN2...More data for this Ligand-Target Pair
TargetGlutamate receptor ionotropic, NMDA 2A(Homo sapiens (Human))
Takeda Pharmaceutical
Curated by ChEMBL
Takeda Pharmaceutical
Curated by ChEMBL
Affinity DataEC50: 147nMAssay Description:The whole-cell patch-clamp technique was used to investigate the effects of positive allosteric modulating activity of test compounds on GlunN1/GluN2...More data for this Ligand-Target Pair
TargetGlutamate receptor ionotropic, NMDA 2A(Homo sapiens (Human))
Takeda Pharmaceutical
Curated by ChEMBL
Takeda Pharmaceutical
Curated by ChEMBL
Affinity DataEC50: 148nMAssay Description:The whole-cell patch-clamp technique was used to investigate the effects of positive allosteric modulating activity of test compounds on GlunN1/GluN2...More data for this Ligand-Target Pair
TargetGlutamate receptor ionotropic, NMDA 2A(Homo sapiens (Human))
Takeda Pharmaceutical
Curated by ChEMBL
Takeda Pharmaceutical
Curated by ChEMBL
Affinity DataEC50: 150nMAssay Description:Positive allosteric modulator activity at GluN2A NMDA receptor (unknown origin) expressed in CHO cells co-expressing GluN1a in the presence of L-glut...More data for this Ligand-Target Pair
Ligand Info
TargetGlutamate receptor ionotropic, NMDA 2A(Homo sapiens (Human))
Takeda Pharmaceutical
Curated by ChEMBL
Takeda Pharmaceutical
Curated by ChEMBL
Affinity DataEC50: 154nMAssay Description:The whole-cell patch-clamp technique was used to investigate the effects of positive allosteric modulating activity of test compounds on GlunN1/GluN2...More data for this Ligand-Target Pair
TargetGlutamate receptor ionotropic, NMDA 2A(Homo sapiens (Human))
Takeda Pharmaceutical
Curated by ChEMBL
Takeda Pharmaceutical
Curated by ChEMBL
Affinity DataEC50: 158nMAssay Description:The whole-cell patch-clamp technique was used to investigate the effects of positive allosteric modulating activity of test compounds on GlunN1/GluN2...More data for this Ligand-Target Pair
TargetGlutamate receptor ionotropic, NMDA 2A(Homo sapiens (Human))
Takeda Pharmaceutical
Curated by ChEMBL
Takeda Pharmaceutical
Curated by ChEMBL
Affinity DataEC50: 163nMAssay Description:The whole-cell patch-clamp technique was used to investigate the effects of positive allosteric modulating activity of test compounds on GlunN1/GluN2...More data for this Ligand-Target Pair
TargetGlutamate receptor ionotropic, NMDA 2A(Homo sapiens (Human))
Takeda Pharmaceutical
Curated by ChEMBL
Takeda Pharmaceutical
Curated by ChEMBL
Affinity DataEC50: 165nMAssay Description:The whole-cell patch-clamp technique was used to investigate the effects of positive allosteric modulating activity of test compounds on GlunN1/GluN2...More data for this Ligand-Target Pair
TargetGlutamate receptor ionotropic, NMDA 2A(Homo sapiens (Human))
Takeda Pharmaceutical
Curated by ChEMBL
Takeda Pharmaceutical
Curated by ChEMBL
Affinity DataEC50: 165nMAssay Description:The whole-cell patch-clamp technique was used to investigate the effects of positive allosteric modulating activity of test compounds on GlunN1/GluN2...More data for this Ligand-Target Pair
TargetGlutamate receptor ionotropic, NMDA 2A(Homo sapiens (Human))
Takeda Pharmaceutical
Curated by ChEMBL
Takeda Pharmaceutical
Curated by ChEMBL
Affinity DataEC50: 165nMAssay Description:The whole-cell patch-clamp technique was used to investigate the effects of positive allosteric modulating activity of test compounds on GlunN1/GluN2...More data for this Ligand-Target Pair
TargetGlutamate receptor ionotropic, NMDA 2A(Homo sapiens (Human))
Takeda Pharmaceutical
Curated by ChEMBL
Takeda Pharmaceutical
Curated by ChEMBL
Affinity DataEC50: 169nMAssay Description:The whole-cell patch-clamp technique was used to investigate the effects of positive allosteric modulating activity of test compounds on GlunN1/GluN2...More data for this Ligand-Target Pair