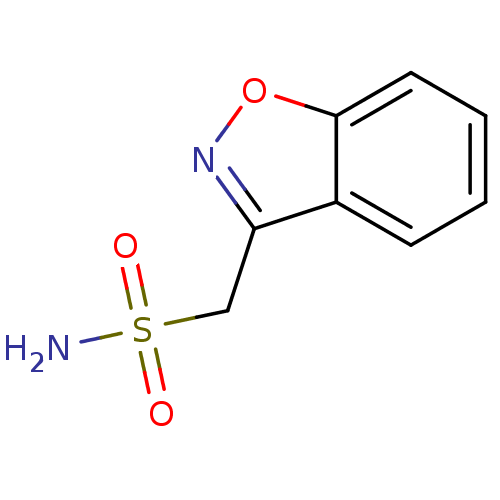
Report error Found 18 Enz. Inhib. hit(s) with all data for entry = 8235
Affinity DataKi: 11nM ΔG°: -45.4kJ/molepH: 7.5 T: 2°CAssay Description:Phenol red (at a concentration of 0.2 mM) has been used as indicator, working at the absorbance maximum of 557 nm, with 10 mM Tris-HCl (pH 7.5) as bu...More data for this Ligand-Target Pair
Affinity DataKi: 15nM ΔG°: -44.7kJ/molepH: 7.5 T: 2°CAssay Description:Phenol red (at a concentration of 0.2 mM) has been used as indicator, working at the absorbance maximum of 557 nm, with 10 mM Tris-HCl (pH 7.5) as bu...More data for this Ligand-Target Pair
Affinity DataKi: 15nM ΔG°: -44.7kJ/molepH: 7.5 T: 2°CAssay Description:Phenol red (at a concentration of 0.2 mM) has been used as indicator, working at the absorbance maximum of 557 nm, with 10 mM Tris-HCl (pH 7.5) as bu...More data for this Ligand-Target Pair
Affinity DataKi: 30nM ΔG°: -42.9kJ/molepH: 7.5 T: 2°CAssay Description:Phenol red (at a concentration of 0.2 mM) has been used as indicator, working at the absorbance maximum of 557 nm, with 10 mM Tris-HCl (pH 7.5) as bu...More data for this Ligand-Target Pair
Affinity DataKi: 41nM ΔG°: -42.2kJ/molepH: 7.5 T: 2°CAssay Description:Phenol red (at a concentration of 0.2 mM) has been used as indicator, working at the absorbance maximum of 557 nm, with 10 mM Tris-HCl (pH 7.5) as bu...More data for this Ligand-Target Pair
Affinity DataKi: 50nM ΔG°: -41.7kJ/molepH: 7.5 T: 2°CAssay Description:Phenol red (at a concentration of 0.2 mM) has been used as indicator, working at the absorbance maximum of 557 nm, with 10 mM Tris-HCl (pH 7.5) as bu...More data for this Ligand-Target Pair
Affinity DataKi: 60nM ΔG°: -41.2kJ/molepH: 7.5 T: 2°CAssay Description:Phenol red (at a concentration of 0.2 mM) has been used as indicator, working at the absorbance maximum of 557 nm, with 10 mM Tris-HCl (pH 7.5) as bu...More data for this Ligand-Target Pair
Affinity DataKi: 62nM ΔG°: -41.1kJ/molepH: 7.5 T: 2°CAssay Description:Phenol red (at a concentration of 0.2 mM) has been used as indicator, working at the absorbance maximum of 557 nm, with 10 mM Tris-HCl (pH 7.5) as bu...More data for this Ligand-Target Pair
Affinity DataKi: 85nM ΔG°: -40.4kJ/molepH: 7.5 T: 2°CAssay Description:Phenol red (at a concentration of 0.2 mM) has been used as indicator, working at the absorbance maximum of 557 nm, with 10 mM Tris-HCl (pH 7.5) as bu...More data for this Ligand-Target Pair
Affinity DataKi: 95nM ΔG°: -40.1kJ/molepH: 7.5 T: 2°CAssay Description:Phenol red (at a concentration of 0.2 mM) has been used as indicator, working at the absorbance maximum of 557 nm, with 10 mM Tris-HCl (pH 7.5) as bu...More data for this Ligand-Target Pair
Affinity DataKi: 105nM ΔG°: -39.8kJ/molepH: 7.5 T: 2°CAssay Description:Phenol red (at a concentration of 0.2 mM) has been used as indicator, working at the absorbance maximum of 557 nm, with 10 mM Tris-HCl (pH 7.5) as bu...More data for this Ligand-Target Pair
Affinity DataKi: 128nM ΔG°: -39.3kJ/molepH: 7.5 T: 2°CAssay Description:Phenol red (at a concentration of 0.2 mM) has been used as indicator, working at the absorbance maximum of 557 nm, with 10 mM Tris-HCl (pH 7.5) as bu...More data for this Ligand-Target Pair
Affinity DataKi: 270nM ΔG°: -37.5kJ/molepH: 7.5 T: 2°CAssay Description:Phenol red (at a concentration of 0.2 mM) has been used as indicator, working at the absorbance maximum of 557 nm, with 10 mM Tris-HCl (pH 7.5) as bu...More data for this Ligand-Target Pair
Affinity DataKi: 380nM ΔG°: -36.6kJ/molepH: 7.5 T: 2°CAssay Description:Phenol red (at a concentration of 0.2 mM) has been used as indicator, working at the absorbance maximum of 557 nm, with 10 mM Tris-HCl (pH 7.5) as bu...More data for this Ligand-Target Pair
Affinity DataKi: 593nM ΔG°: -35.5kJ/molepH: 7.5 T: 2°CAssay Description:Phenol red (at a concentration of 0.2 mM) has been used as indicator, working at the absorbance maximum of 557 nm, with 10 mM Tris-HCl (pH 7.5) as bu...More data for this Ligand-Target Pair
Affinity DataKi: 784nM ΔG°: -34.8kJ/molepH: 7.5 T: 2°CAssay Description:Phenol red (at a concentration of 0.2 mM) has been used as indicator, working at the absorbance maximum of 557 nm, with 10 mM Tris-HCl (pH 7.5) as bu...More data for this Ligand-Target Pair
Affinity DataKi: 1.15E+3nM ΔG°: -33.9kJ/molepH: 7.5 T: 2°CAssay Description:Phenol red (at a concentration of 0.2 mM) has been used as indicator, working at the absorbance maximum of 557 nm, with 10 mM Tris-HCl (pH 7.5) as bu...More data for this Ligand-Target Pair
Affinity DataKi: 2.27E+3nM ΔG°: -32.2kJ/molepH: 7.5 T: 2°CAssay Description:Phenol red (at a concentration of 0.2 mM) has been used as indicator, working at the absorbance maximum of 557 nm, with 10 mM Tris-HCl (pH 7.5) as bu...More data for this Ligand-Target Pair
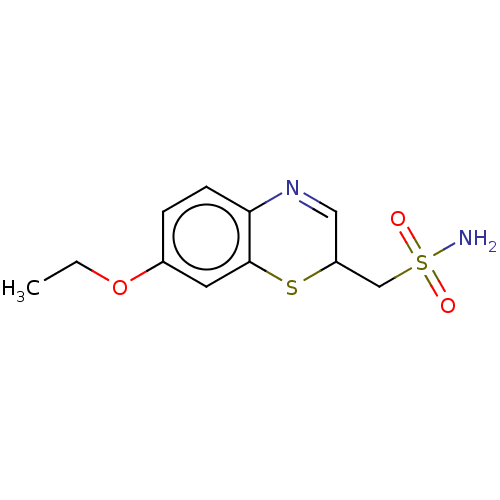
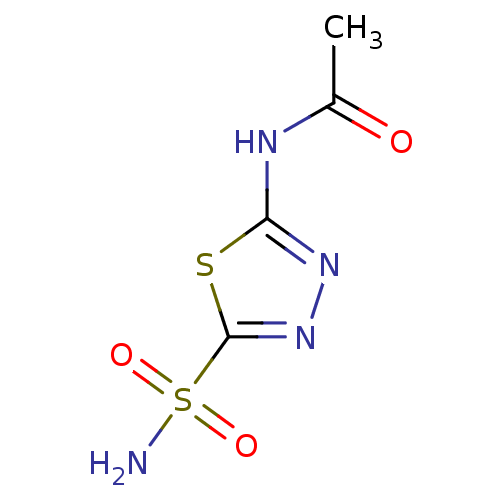
 3D Structure (crystal)
3D Structure (crystal)