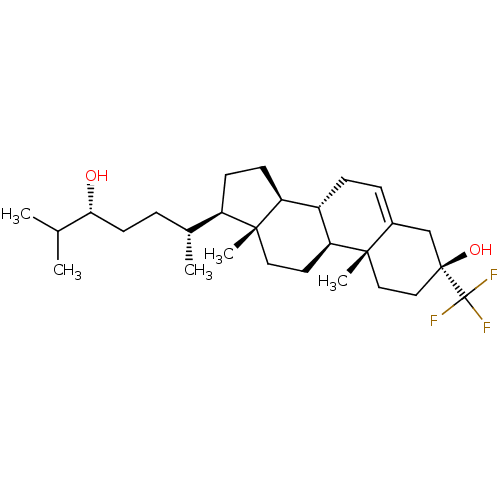
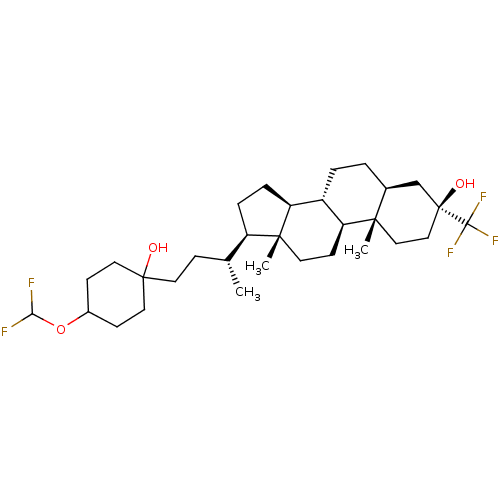
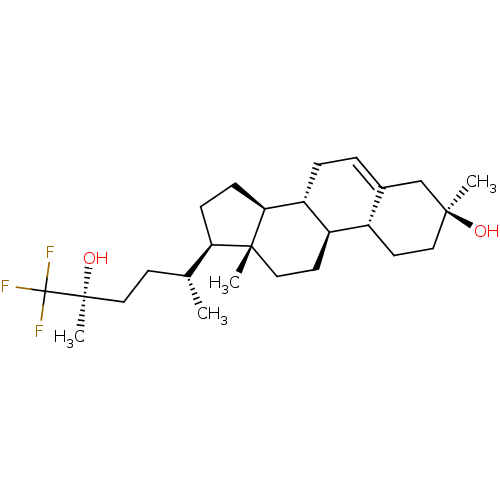
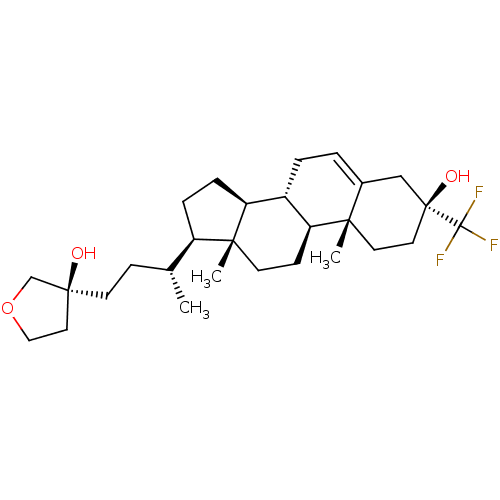
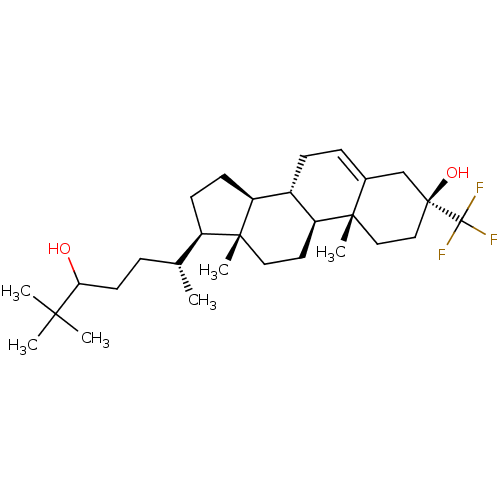
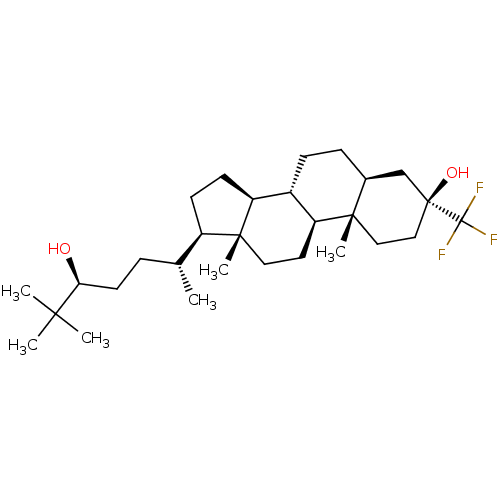
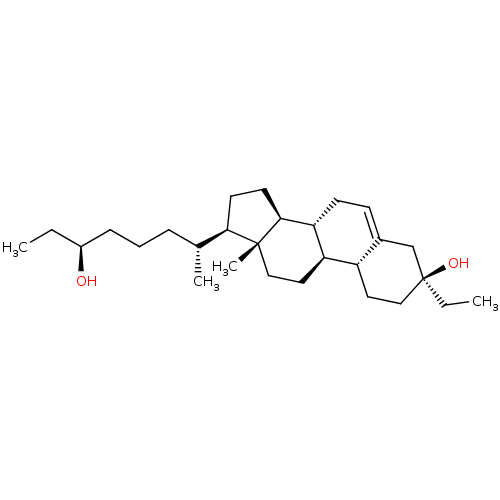
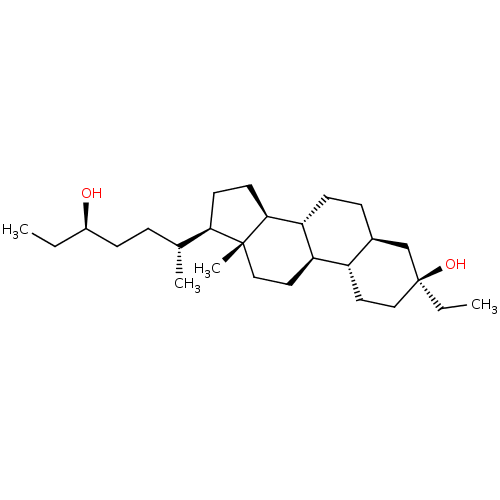
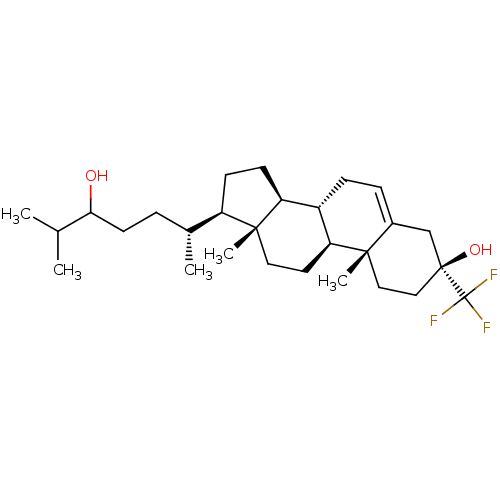
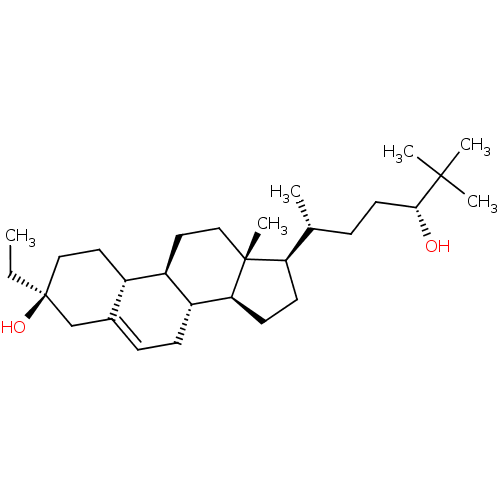
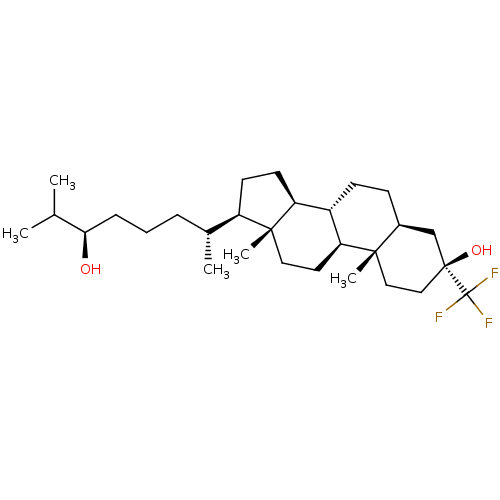
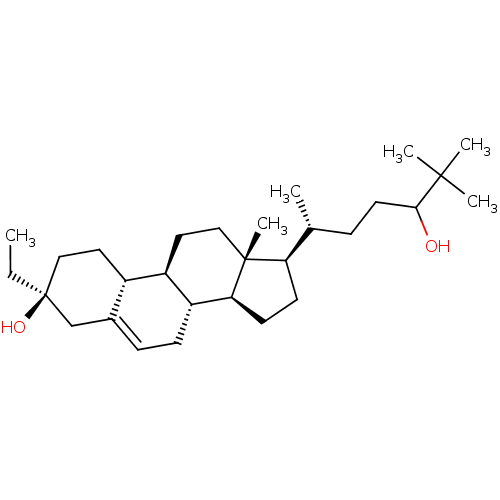
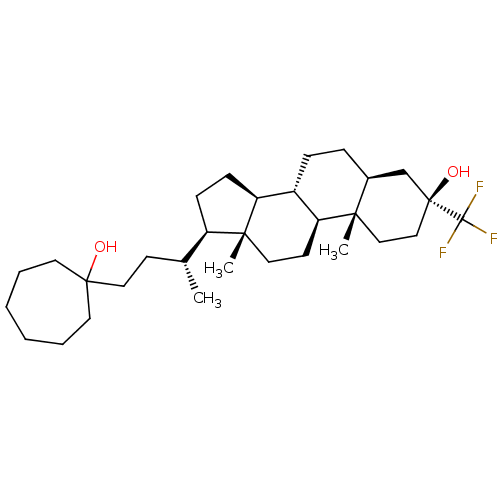
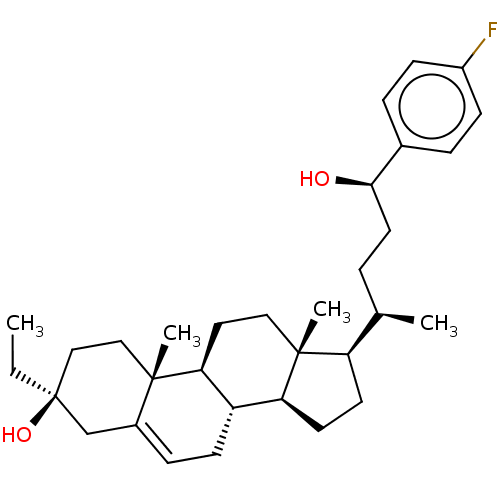
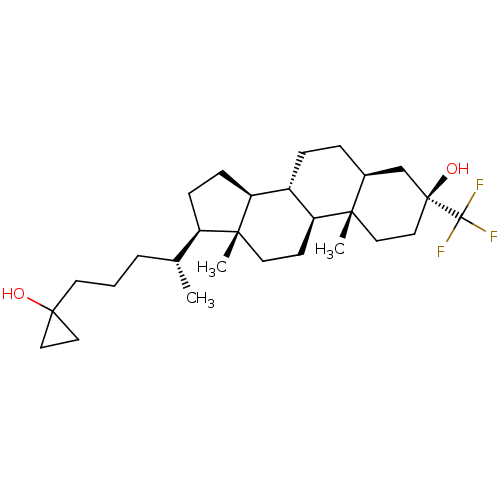
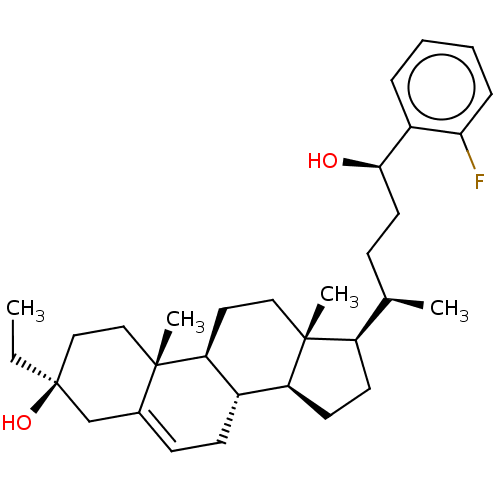
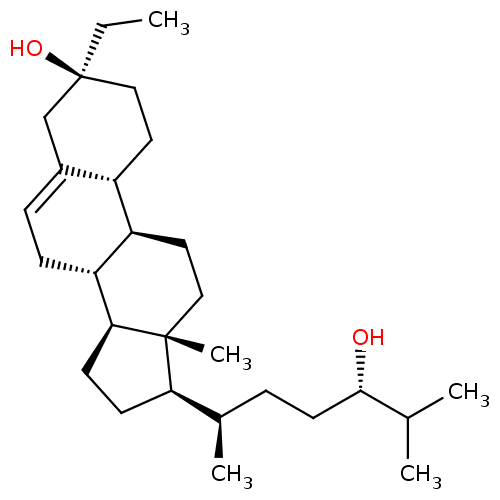
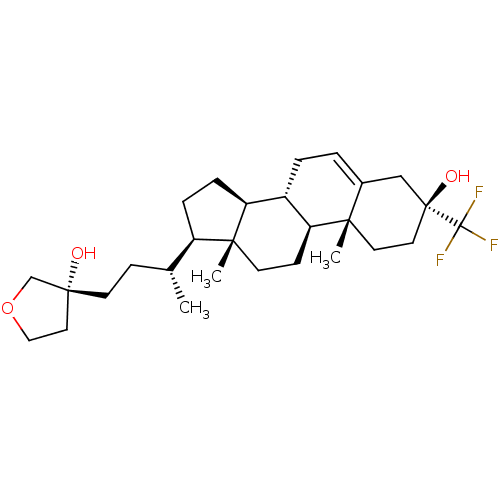
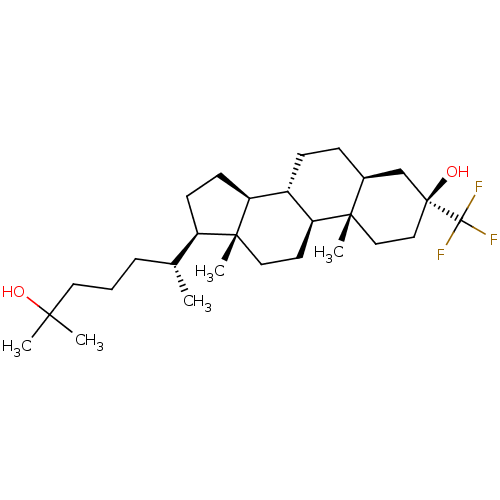
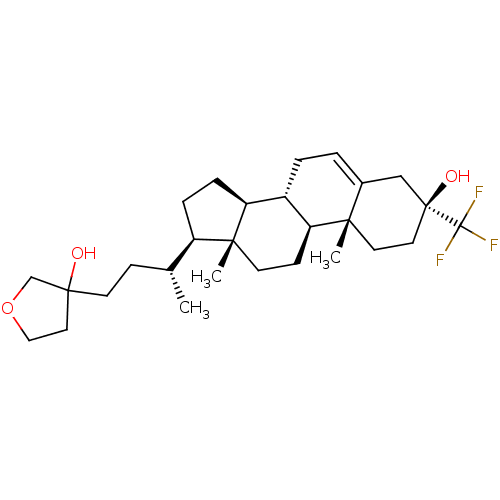
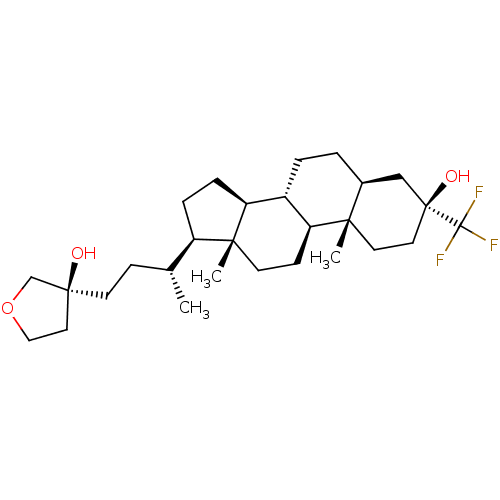
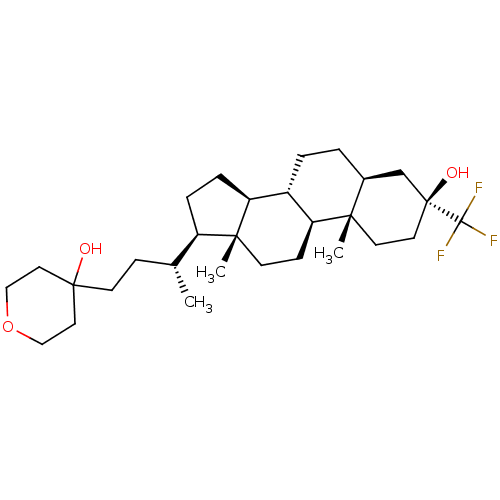
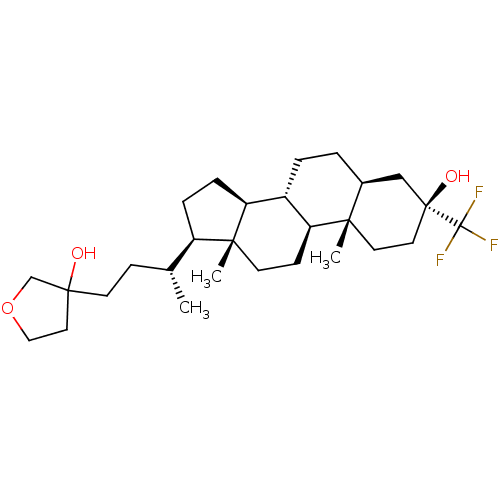
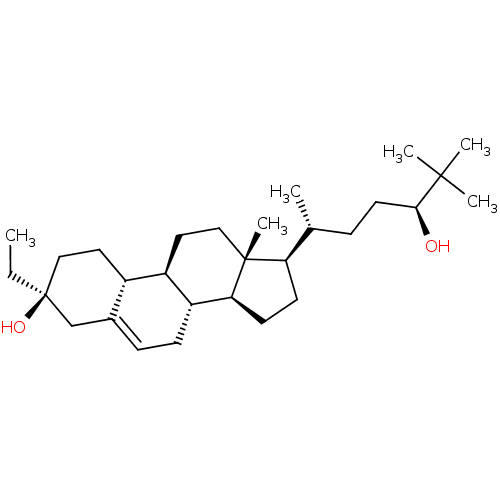
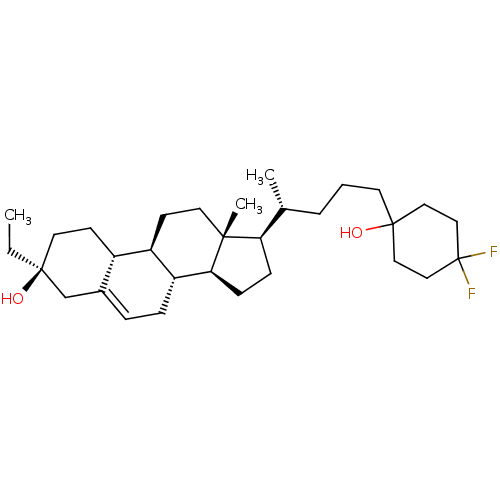
Report error Found 314 Enz. Inhib. hit(s) with all data for entry = 10229
Affinity DataEC50: 31.6nMAssay Description:The whole-cell patch-clamp technique was used to investigate the effects of positive allosteric modulating activity of test compounds on GlunN1/GluN2...More data for this Ligand-Target Pair
Affinity DataEC50: 33.6nMAssay Description:The whole-cell patch-clamp technique was used to investigate the effects of positive allosteric modulating activity of test compounds on GlunN1/GluN2...More data for this Ligand-Target Pair
Affinity DataEC50: 33.6nMAssay Description:The whole-cell patch-clamp technique was used to investigate the effects of positive allosteric modulating activity of test compounds on GlunN1/GluN2...More data for this Ligand-Target Pair
Affinity DataEC50: 34.5nMAssay Description:The whole-cell patch-clamp technique was used to investigate the effects of positive allosteric modulating activity of test compounds on GlunN1/GluN2...More data for this Ligand-Target Pair
Affinity DataEC50: 35.1nMAssay Description:The whole-cell patch-clamp technique was used to investigate the effects of positive allosteric modulating activity of test compounds on GlunN1/GluN2...More data for this Ligand-Target Pair
Affinity DataEC50: 47.5nMAssay Description:The whole-cell patch-clamp technique was used to investigate the effects of positive allosteric modulating activity of test compounds on GlunN1/GluN2...More data for this Ligand-Target Pair
Affinity DataEC50: 47.6nMAssay Description:The whole-cell patch-clamp technique was used to investigate the effects of positive allosteric modulating activity of test compounds on GlunN1/GluN2...More data for this Ligand-Target Pair
Affinity DataEC50: 50.1nMAssay Description:The whole-cell patch-clamp technique was used to investigate the effects of positive allosteric modulating activity of test compounds on GlunN1/GluN2...More data for this Ligand-Target Pair
Affinity DataEC50: 53.4nMAssay Description:The whole-cell patch-clamp technique was used to investigate the effects of positive allosteric modulating activity of test compounds on GlunN1/GluN2...More data for this Ligand-Target Pair
Affinity DataEC50: 54.2nMAssay Description:The whole-cell patch-clamp technique was used to investigate the effects of positive allosteric modulating activity of test compounds on GlunN1/GluN2...More data for this Ligand-Target Pair
Affinity DataEC50: 60.7nMAssay Description:The whole-cell patch-clamp technique was used to investigate the effects of positive allosteric modulating activity of test compounds on GlunN1/GluN2...More data for this Ligand-Target Pair
Affinity DataEC50: 61nMAssay Description:The whole-cell patch-clamp technique was used to investigate the effects of positive allosteric modulating activity of test compounds on GlunN1/GluN2...More data for this Ligand-Target Pair
Affinity DataEC50: 61.4nMAssay Description:The whole-cell patch-clamp technique was used to investigate the effects of positive allosteric modulating activity of test compounds on GlunN1/GluN2...More data for this Ligand-Target Pair
Affinity DataEC50: 63.3nMAssay Description:The whole-cell patch-clamp technique was used to investigate the effects of positive allosteric modulating activity of test compounds on GlunN1/GluN2...More data for this Ligand-Target Pair
Affinity DataEC50: 65.8nMAssay Description:The whole-cell patch-clamp technique was used to investigate the effects of positive allosteric modulating activity of test compounds on GlunN1/GluN2...More data for this Ligand-Target Pair
Affinity DataEC50: 66.4nMAssay Description:The whole-cell patch-clamp technique was used to investigate the effects of positive allosteric modulating activity of test compounds on GlunN1/GluN2...More data for this Ligand-Target Pair
Affinity DataEC50: 67.6nMAssay Description:The whole-cell patch-clamp technique was used to investigate the effects of positive allosteric modulating activity of test compounds on GlunN1/GluN2...More data for this Ligand-Target Pair
Affinity DataEC50: 69.7nMAssay Description:The whole-cell patch-clamp technique was used to investigate the effects of positive allosteric modulating activity of test compounds on GlunN1/GluN2...More data for this Ligand-Target Pair
Affinity DataEC50: 70.4nMAssay Description:The whole-cell patch-clamp technique was used to investigate the effects of positive allosteric modulating activity of test compounds on GlunN1/GluN2...More data for this Ligand-Target Pair
Affinity DataEC50: 70.9nMAssay Description:The whole-cell patch-clamp technique was used to investigate the effects of positive allosteric modulating activity of test compounds on GlunN1/GluN2...More data for this Ligand-Target Pair
Affinity DataEC50: 73.4nMAssay Description:The whole-cell patch-clamp technique was used to investigate the effects of positive allosteric modulating activity of test compounds on GlunN1/GluN2...More data for this Ligand-Target Pair
Affinity DataEC50: 75.1nMAssay Description:The whole-cell patch-clamp technique was used to investigate the effects of positive allosteric modulating activity of test compounds on GlunN1/GluN2...More data for this Ligand-Target Pair
Affinity DataEC50: 75.7nMAssay Description:The whole-cell patch-clamp technique was used to investigate the effects of positive allosteric modulating activity of test compounds on GlunN1/GluN2...More data for this Ligand-Target Pair
Affinity DataEC50: 76.9nMAssay Description:The whole-cell patch-clamp technique was used to investigate the effects of positive allosteric modulating activity of test compounds on GlunN1/GluN2...More data for this Ligand-Target Pair
Affinity DataEC50: 78.1nMAssay Description:The whole-cell patch-clamp technique was used to investigate the effects of positive allosteric modulating activity of test compounds on GlunN1/GluN2...More data for this Ligand-Target Pair
Affinity DataEC50: 80nMAssay Description:The whole-cell patch-clamp technique was used to investigate the effects of positive allosteric modulating activity of test compounds on GlunN1/GluN2...More data for this Ligand-Target Pair
Affinity DataEC50: 81.2nMAssay Description:The whole-cell patch-clamp technique was used to investigate the effects of positive allosteric modulating activity of test compounds on GlunN1/GluN2...More data for this Ligand-Target Pair
Affinity DataEC50: 85.1nMAssay Description:The whole-cell patch-clamp technique was used to investigate the effects of positive allosteric modulating activity of test compounds on GlunN1/GluN2...More data for this Ligand-Target Pair
Affinity DataEC50: 87nMAssay Description:The whole-cell patch-clamp technique was used to investigate the effects of positive allosteric modulating activity of test compounds on GlunN1/GluN2...More data for this Ligand-Target Pair
Affinity DataEC50: 88.1nMAssay Description:The whole-cell patch-clamp technique was used to investigate the effects of positive allosteric modulating activity of test compounds on GlunN1/GluN2...More data for this Ligand-Target Pair
Affinity DataEC50: 91.1nMAssay Description:The whole-cell patch-clamp technique was used to investigate the effects of positive allosteric modulating activity of test compounds on GlunN1/GluN2...More data for this Ligand-Target Pair
Affinity DataEC50: 91.7nMAssay Description:The whole-cell patch-clamp technique was used to investigate the effects of positive allosteric modulating activity of test compounds on GlunN1/GluN2...More data for this Ligand-Target Pair
Affinity DataEC50: 91.9nMAssay Description:The whole-cell patch-clamp technique was used to investigate the effects of positive allosteric modulating activity of test compounds on GlunN1/GluN2...More data for this Ligand-Target Pair
Affinity DataEC50: 95.9nMAssay Description:The whole-cell patch-clamp technique was used to investigate the effects of positive allosteric modulating activity of test compounds on GlunN1/GluN2...More data for this Ligand-Target Pair
Affinity DataEC50: 96.2nMAssay Description:The whole-cell patch-clamp technique was used to investigate the effects of positive allosteric modulating activity of test compounds on GlunN1/GluN2...More data for this Ligand-Target Pair
Affinity DataEC50: 96.7nMAssay Description:The whole-cell patch-clamp technique was used to investigate the effects of positive allosteric modulating activity of test compounds on GlunN1/GluN2...More data for this Ligand-Target Pair
Affinity DataEC50: 102nMAssay Description:The whole-cell patch-clamp technique was used to investigate the effects of positive allosteric modulating activity of test compounds on GlunN1/GluN2...More data for this Ligand-Target Pair
Affinity DataEC50: 102nMAssay Description:The whole-cell patch-clamp technique was used to investigate the effects of positive allosteric modulating activity of test compounds on GlunN1/GluN2...More data for this Ligand-Target Pair
Affinity DataEC50: 102nMAssay Description:The whole-cell patch-clamp technique was used to investigate the effects of positive allosteric modulating activity of test compounds on GlunN1/GluN2...More data for this Ligand-Target Pair
Affinity DataEC50: 103nMAssay Description:The whole-cell patch-clamp technique was used to investigate the effects of positive allosteric modulating activity of test compounds on GlunN1/GluN2...More data for this Ligand-Target Pair
Affinity DataEC50: 104nMAssay Description:The whole-cell patch-clamp technique was used to investigate the effects of positive allosteric modulating activity of test compounds on GlunN1/GluN2...More data for this Ligand-Target Pair
Affinity DataEC50: 104nMAssay Description:The whole-cell patch-clamp technique was used to investigate the effects of positive allosteric modulating activity of test compounds on GlunN1/GluN2...More data for this Ligand-Target Pair
Affinity DataEC50: 109nMAssay Description:The whole-cell patch-clamp technique was used to investigate the effects of positive allosteric modulating activity of test compounds on GlunN1/GluN2...More data for this Ligand-Target Pair
Affinity DataEC50: 113nMAssay Description:The whole-cell patch-clamp technique was used to investigate the effects of positive allosteric modulating activity of test compounds on GlunN1/GluN2...More data for this Ligand-Target Pair
Affinity DataEC50: 114nMAssay Description:The whole-cell patch-clamp technique was used to investigate the effects of positive allosteric modulating activity of test compounds on GlunN1/GluN2...More data for this Ligand-Target Pair
Affinity DataEC50: 117nMAssay Description:The whole-cell patch-clamp technique was used to investigate the effects of positive allosteric modulating activity of test compounds on GlunN1/GluN2...More data for this Ligand-Target Pair
Affinity DataEC50: 119nMAssay Description:The whole-cell patch-clamp technique was used to investigate the effects of positive allosteric modulating activity of test compounds on GlunN1/GluN2...More data for this Ligand-Target Pair
Affinity DataEC50: 120nMAssay Description:The whole-cell patch-clamp technique was used to investigate the effects of positive allosteric modulating activity of test compounds on GlunN1/GluN2...More data for this Ligand-Target Pair
Affinity DataEC50: 124nMAssay Description:The whole-cell patch-clamp technique was used to investigate the effects of positive allosteric modulating activity of test compounds on GlunN1/GluN2...More data for this Ligand-Target Pair
Affinity DataEC50: 124nMAssay Description:The whole-cell patch-clamp technique was used to investigate the effects of positive allosteric modulating activity of test compounds on GlunN1/GluN2...More data for this Ligand-Target Pair