Reaction Details Report a problem with these data
Report a problem with these data
 Report a problem with these data
Report a problem with these dataTarget
DNA gyrase subunit A/B
Ligand
BDBM50577590
Substrate
n/a
Meas. Tech.
ChEMBL_2130602 (CHEMBL4840031)
IC50
160±n/a nM
Citation
 Lu, Y; Vibhute, S; Li, L; Okumu, A; Ratigan, SC; Nolan, S; Papa, JL; Mann, CA; English, A; Chen, A; Seffernick, JT; Koci, B; Duncan, LR; Roth, B; Cummings, JE; Slayden, RA; Lindert, S; McElroy, CA; Wozniak, DJ; Yalowich, J; Mitton-Fry, MJ Optimization of TopoIV Potency, ADMET Properties, and hERG Inhibition of 5-Amino-1,3-dioxane-Linked Novel Bacterial Topoisomerase Inhibitors: Identification of a Lead with J Med Chem 64:15214-15249 (2021) [PubMed] Article
Lu, Y; Vibhute, S; Li, L; Okumu, A; Ratigan, SC; Nolan, S; Papa, JL; Mann, CA; English, A; Chen, A; Seffernick, JT; Koci, B; Duncan, LR; Roth, B; Cummings, JE; Slayden, RA; Lindert, S; McElroy, CA; Wozniak, DJ; Yalowich, J; Mitton-Fry, MJ Optimization of TopoIV Potency, ADMET Properties, and hERG Inhibition of 5-Amino-1,3-dioxane-Linked Novel Bacterial Topoisomerase Inhibitors: Identification of a Lead with J Med Chem 64:15214-15249 (2021) [PubMed] Article More Info.:
Target
Name:
DNA gyrase subunit A/B
Synonyms:
DNA Gyrase
Type:
A2B2 tetramer
Mol. Mass.:
n/a
Description:
n/a
Components:
This complex has 2 components.
Component 1
Name:
DNA gyrase subunit A
Synonyms:
DNA gyrase subunit A (gyrA) | GYRA_STAAU | gyrA
Type:
Enzyme Subunit
Mol. Mass.:
99588.82
Organism:
Staphylococcus aureus
Description:
n/a
Residue:
889
Sequence:
MAELPQSRINERNITSEMRESFLDYAMSVIVARALPDVRDGLKPVHRRILYGLNEQGMTPDKSYKKSARIVGDVMGKYHPHGDSSIYEAMVRMAQDFSYRYPLVDGQGNFGSMDGDGAAAMRYTEARMTKITLELLRDINKDTIDFIDNYDGNEREPSVLPARFPNLLANGASGIAVGMATNIPPHNLTELINGVLSLSKNPDISIAELMEDIEGPDFPTAGLILGKSGIRRAYETGRGSIQMRSRAVIEERGGGRQRIVVTEIPFQVNKARMIEKIAELVRDKKIDGITDLRDETSLRTGVRVVIDVRKDANASVILNNLYKQTPLQTSFGVNMIALVNGRPKLINLKEALVHYLEHQKTVVRRRTQYNLRKAKDRAHILEGLRIALDHIDEIISTIRESDTDKVAMESLQQRFKLSEKQAQAILDMRLRRLTGLERDKIEAEYNELLNYISELEAILADEEVLLQLVRDELTEIRDRFGDDRRTEIQLGGFEDLEDEDLIPEEQIVITLSHNNYIKRLPVSTYRAQNRGGRGVQGMNTLEEDFVSQLVTLSTHDHVLFFTNKGRVYKLKGYEVPELSRQSKGIPVVNAIELENDEVISTMIAVKDLESEDNFLVFATKRGVVKRSALSNFSRINRNGKIAISFREDDELIAVRLTSGQEDILIGTSHASLIRFPESTLRPLGRTATGVKGITLREGDEVVGLDVAHANSVDEVLVVTENGYGKRTPVNDYRLSNRGGKGIKTATITERNGNVVCITTVTGEEDLMIVTNAGVIIRLDVADISQNGRAAQGVRLIRLGDDQFVSTVAKVKEDAEDETNEDEQSTSTVSEDGTEQQREAVVNDETPGNAIHTEVIDSEENDEDGRIEVRQDFMDRVEEDIQQSLDEDEE
Component 2
Name:
DNA gyrase subunit B
Synonyms:
DNA gyrase | DNA gyrase subunit B (DNA gyraseB) | DNA gyrase subunit B (gyrB) | GYRB_STAAU | gyrB
Type:
Enzyme Subunit
Mol. Mass.:
72530.91
Organism:
Staphylococcus aureus
Description:
n/a
Residue:
644
Sequence:
MVTALSDVNNTDNYGAGQIQVLEGLEAVRKRPGMYIGSTSERGLHHLVWEIVDNSIDEALAGYANQIEVVIEKDNWIKVTDNGRGIPVDIQEKMGRPAVEVILTVLHAGGKFGGGGYKVSGGLHGVGSSVVNALSQDLEVYVHRNETIYHQAYKKGVPQFDLKEVGTTDKTGTVIRFKADGEIFTETTVYNYETLQQRIRELAFLNKGIQITLRDERDEENVREDSYHYEGGIKSYVELLNENKEPIHDEPIYIHQSKDDIEVEIAIQYNSGYATNLLTYANNIHTYEGGTHEDGFKRALTRVLNSYGLSSKIMKEEKDRLSGEDTREGMTAIISIKHGDPQFEGQTKTKLGNSEVRQVVDKLFSEHFERFLYENPQVARTVVEKGIMAARARVAAKKAREVTRRKSALDVASLPGKLADCSSKSPEECEIFLVEGDSAGGSTKSGRDSRTQAILPLRGKILNVEKARLDRILNNNEIRQMITAFGTGIGGDFDLAKARYHKIVIMTDADVDGAHIRTLLLTFFYRFMRPLIEAGYVYIAQPPLYKLTQGKQKYYVYNDRELDKLKSELNPTPKWSIARYKGLGEMNADQLWETTMNPEHRALLQVKLEDAIEADQTFEMLMGDVVENRRQFIEDNAVYANLDF
Inhibitor
Name:
BDBM50577590
Synonyms:
CHEMBL4868523
Type:
Small organic molecule
Emp. Form.:
C23H26FN3O4
Mol. Mass.:
427.4686
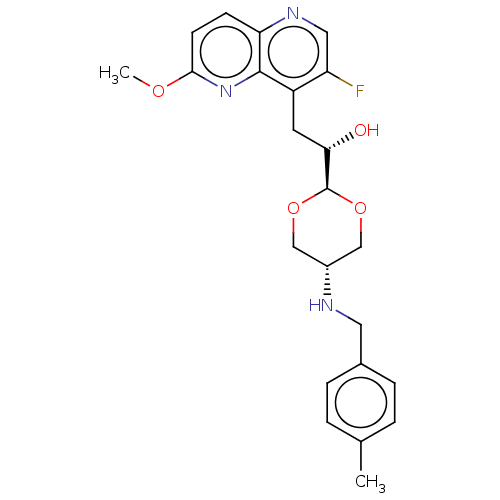
SMILES:
[H][C@@]1(OC[C@@H](CO1)NCc1ccc(C)cc1)[C@@H](O)Cc1c(F)cnc2ccc(OC)nc12 |r,wU:16.18,wD:4.7,1.0,(67.04,-52.95,;65.71,-52.19,;64.37,-51.44,;64.37,-49.89,;65.7,-49.12,;67.04,-49.89,;67.04,-51.42,;65.7,-47.58,;67.03,-46.8,;68.37,-47.57,;68.37,-49.11,;69.7,-49.88,;71.03,-49.11,;72.37,-49.87,;71.03,-47.56,;69.69,-46.8,;65.71,-53.73,;67.06,-54.49,;64.4,-54.5,;64.4,-56.04,;65.73,-56.8,;67.06,-56.03,;65.73,-58.35,;64.41,-59.13,;63.07,-58.36,;61.74,-59.14,;60.41,-58.37,;60.41,-56.82,;59.07,-56.05,;59.07,-54.51,;61.74,-56.05,;63.07,-56.82,)|