Reaction Details Report a problem with these data
Report a problem with these data
 Report a problem with these data
Report a problem with these dataTarget
Neuronal acetylcholine receptor subunit alpha-7
Ligand
BDBM50296733
Substrate
n/a
Meas. Tech.
ChEMBL_582484 (CHEMBL1058161)
Ki
749±n/a nM
Citation
 O'Donnell, CJ; Peng, L; O'Neill, BT; Arnold, EP; Mather, RJ; Sands, SB; Shrikhande, A; Lebel, LA; Spracklin, DK; Nedza, FM Synthesis and SAR studies of 1,4-diazabicyclo[3.2.2]nonane phenyl carbamates--subtype selective, high affinity alpha7 nicotinic acetylcholine receptor agonists. Bioorg Med Chem Lett 19:4747-51 (2009) [PubMed] Article
O'Donnell, CJ; Peng, L; O'Neill, BT; Arnold, EP; Mather, RJ; Sands, SB; Shrikhande, A; Lebel, LA; Spracklin, DK; Nedza, FM Synthesis and SAR studies of 1,4-diazabicyclo[3.2.2]nonane phenyl carbamates--subtype selective, high affinity alpha7 nicotinic acetylcholine receptor agonists. Bioorg Med Chem Lett 19:4747-51 (2009) [PubMed] Article More Info.:
Target
Name:
Neuronal acetylcholine receptor subunit alpha-7
Synonyms:
ACHA7_RAT | Acra7 | Cholinergic, Nicotinic Alpha7 | Cholinergic, Nicotinic Alpha7/5-HT3 | Chrna7 | Neuronal acetylcholine receptor | Neuronal acetylcholine receptor (alpha7 nAChR) | Neuronal acetylcholine receptor subunit alpha 7 | Neuronal acetylcholine receptor subunit alpha-7 | Neuronal acetylcholine receptor subunit alpha-7 (nAChR alpha7) | Neuronal acetylcholine receptor subunit alpha-7 (nAChR)
Type:
Enzyme
Mol. Mass.:
56502.44
Organism:
Rattus norvegicus (Rat)
Description:
Q05941
Residue:
502
Sequence:
MCGGRGGIWLALAAALLHVSLQGEFQRRLYKELVKNYNPLERPVANDSQPLTVYFSLSLLQIMDVDEKNQVLTTNIWLQMSWTDHYLQWNMSEYPGVKNVRFPDGQIWKPDILLYNSADERFDATFHTNVLVNASGHCQYLPPGIFKSSCYIDVRWFPFDVQQCKLKFGSWSYGGWSLDLQMQEADISSYIPNGEWDLMGIPGKRNEKFYECCKEPYPDVTYTVTMRRRTLYYGLNLLIPCVLISALALLVFLLPADSGEKISLGITVLLSLTVFMLLVAEIMPATSDSVPLIAQYFASTMIIVGLSVVVTVIVLRYHHHDPDGGKMPKWTRIILLNWCAWFLRMKRPGEDKVRPACQHKPRRCSLASVELSAGAGPPTSNGNLLYIGFRGLEGMHCAPTPDSGVVCGRLACSPTHDEHLMHGAHPSDGDPDLAKILEEVRYIANRFRCQDESEVICSEWKFAACVVDRLCLMAFSVFTIICTIGILMSAPNFVEAVSKDFA
Inhibitor
Name:
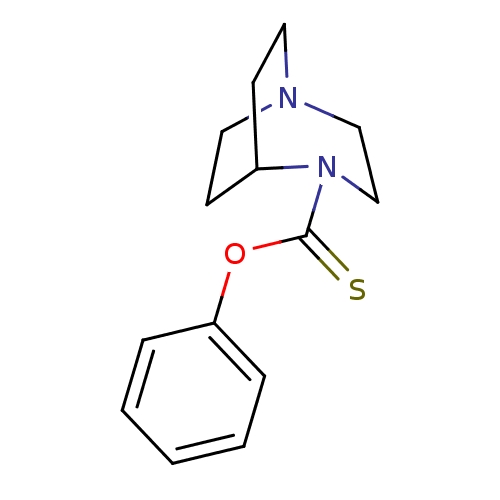
BDBM50296733
Synonyms:
CHEMBL560069 | O-phenyl 1,4-diazabicyclo[3.2.2]nonane-4-carbothioate
Type:
Small organic molecule
Emp. Form.:
C14H18N2OS
Mol. Mass.:
262.371
SMILES:
S=C(Oc1ccccc1)N1CCN2CCC1CC2 |TLB:1:9:14.13:16.17,(30.03,-23.84,;30.3,-25.36,;31.76,-25.89,;32.94,-24.89,;34.38,-25.42,;35.57,-24.43,;35.3,-22.91,;33.83,-22.38,;32.66,-23.38,;29.12,-26.36,;29.49,-27.84,;28.48,-27.11,;27.1,-27.18,;27.02,-25.47,;27.64,-24.32,;27.71,-25.73,;26.4,-26.44,;26.11,-27.89,)|