Reaction Details Report a problem with these data
Report a problem with these data
 Report a problem with these data
Report a problem with these dataTarget
Histone-lysine N-methyltransferase EZH2 [Y641F]
Ligand
BDBM516861
Substrate
n/a
Meas. Tech.
In Vitro Methyltransferase Activity Assay
IC50
12.0±n/a nM
Citation
More Info.:
Target
Name:
Histone-lysine N-methyltransferase EZH2 [Y641F]
Synonyms:
EZH2 | EZH2(Y641F) | EZH2_HUMAN | Histone-lysine N-methyltransferase EZH2 (Y641F) | KMT6
Type:
n/a
Mol. Mass.:
85351.84
Organism:
Homo sapiens (Human)
Description:
Q15910[Y641F]
Residue:
746
Sequence:
MGQTGKKSEKGPVCWRKRVKSEYMRLRQLKRFRRADEVKSMFSSNRQKILERTEILNQEWKQRRIQPVHILTSVSSLRGTRECSVTSDLDFPTQVIPLKTLNAVASVPIMYSWSPLQQNFMVEDETVLHNIPYMGDEVLDQDGTFIEELIKNYDGKVHGDRECGFINDEIFVELVNALGQYNDDDDDDDGDDPEEREEKQKDLEDHRDDKESRPPRKFPSDKIFEAISSMFPDKGTAEELKEKYKELTEQQLPGALPPECTPNIDGPNAKSVQREQSLHSFHTLFCRRCFKYDCFLHPFHATPNTYKRKNTETALDNKPCGPQCYQHLEGAKEFAAALTAERIKTPPKRPGGRRRGRLPNNSSRPSTPTINVLESKDTDSDREAGTETGGENNDKEEEEKKDETSSSSEANSRCQTPIKMKPNIEPPENVEWSGAEASMFRVLIGTYYDNFCAIARLIGTKTCRQVYEFRVKESSIIAPAPAEDVDTPPRKKKRKHRLWAAHCRKIQLKKDGSSNHVYNYQPCDHPRQPCDSSCPCVIAQNFCEKFCQCSSECQNRFPGCRCKAQCNTKQCPCYLAVRECDPDLCLTCGAADHWDSKNVSCKNCSIQRGSKKHLLLAPSDVAGWGIFIKDPVQKNEFISEFCGEIISQDEADRRGKVYDKYMCSFLFNLNNDFVVDATRKGNKIRFANHSVNPNCYAKVMMVNGDHRIGIFAKRAIQTGEELFFDYRYSQADALKYVGIEREMEIP
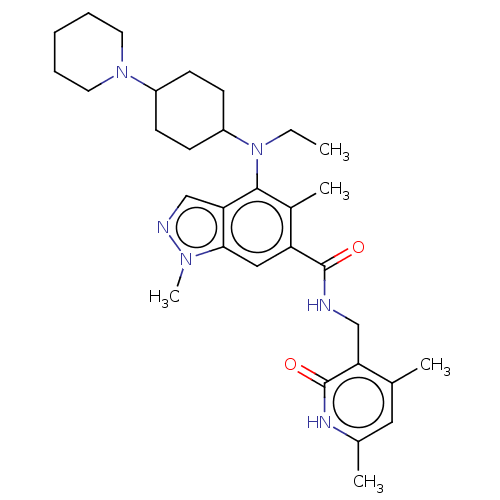
Inhibitor
Name:
BDBM516861
Synonyms:
US11104664, Compound P-32
Type:
Small organic molecule
Emp. Form.:
C31H44N6O2
Mol. Mass.:
532.7201
SMILES:
CCN(C1CCC(CC1)N1CCCCC1)c1c(C)c(cc2n(C)ncc12)C(=O)NCc1c(C)cc(C)[nH]c1=O |(3.33,.77,;1.99,1.54,;.66,.77,;.66,-.77,;-.68,-1.54,;-.68,-3.08,;.66,-3.85,;1.99,-3.08,;1.99,-1.54,;.66,-5.39,;-.68,-6.16,;-.68,-7.7,;.66,-8.47,;1.99,-7.7,;1.99,-6.16,;-.68,1.54,;-.68,3.08,;.66,3.85,;-2.01,3.85,;-3.34,3.08,;-3.34,1.54,;-4.49,.51,;-5.99,.83,;-3.86,-.9,;-2.33,-.74,;-2.01,.77,;-2.01,5.39,;-3.34,6.16,;-.68,6.16,;.66,5.39,;1.99,6.16,;3.33,5.39,;3.33,3.85,;4.66,6.16,;4.66,7.7,;5.99,8.47,;3.33,8.47,;1.99,7.7,;.66,8.47,)|