Reaction Details Report a problem with these data
Report a problem with these data
 Report a problem with these data
Report a problem with these dataTarget
Bcl-2-like protein 11 [51-76]/Induced myeloid leukemia cell differentiation protein Mcl-1 [171-327]
Ligand
BDBM557756
Substrate
n/a
Meas. Tech.
Cell Free Mcl-1: Bim Affinity Assay (Mcl-1 HTRF)
IC50
0.267±n/a nM
Citation
 Brown, SP; Li, K; Li, Y; Reed, AB; Lanman, BA Compounds that inhibit Mcl-1 protein US Patent US11364248 Publication Date 6/21/2022
Brown, SP; Li, K; Li, Y; Reed, AB; Lanman, BA Compounds that inhibit Mcl-1 protein US Patent US11364248 Publication Date 6/21/2022 More Info.:
Target
Name:
Bcl-2-like protein 11 [51-76]/Induced myeloid leukemia cell differentiation protein Mcl-1 [171-327]
Synonyms:
Bcl-2-like protein 11 [51-76]/Induced myeloid leukemia cell differentiation protein Mcl-1 [171-327] | Induced myeloid leukemia cell differentiation protein Mcl-1 [171-327]/Bcl-2-like protein 11 [51-76] | Mcl-1 and BIM | Mcl-1/BIM
Type:
Protein
Mol. Mass.:
n/a
Description:
n/a
Components:
This complex has 2 components.
Component 1
Name:
Induced myeloid leukemia cell differentiation protein Mcl-1 [171-327]
Synonyms:
BCL2L3 | Induced myeloid leukemia cell differentiation protein Mcl-1(171-327) | MCL1 | MCL1_HUMAN | Myeloid cell leukemia 1 (Mcl-1)
Type:
Enzyme Catalytic Domain
Mol. Mass.:
17896.13
Organism:
Homo sapiens (Human)
Description:
Q07820[171-327]
Residue:
157
Sequence:
EDELYRQSLEIISRYLREQATGAKDTKPMGRSGATSRKALETLRRVGDGVQRNHETAFQGMLRKLDIKNEDDVKSLSRVMIHVFSDGVTNWGRIVTLISFGAFVAKHLKTINQESCIEPLAESITDVLVRTKRDWLVKQRGWDGFVEFFHVEDLEGG
Component 2
Name:
Bcl-2-like protein 11 [51-76]
Synonyms:
B2L11_HUMAN | BCL2L11 | BIM | Bcl-2-like protein 11 (51-76) | Bcl-2-like protein 11 (BIM) (51-76)
Type:
Enzyme Catalytic Domain
Mol. Mass.:
2530.22
Organism:
Homo sapiens (Human)
Description:
O43521[51-76]
Residue:
26
Sequence:
EGDSCPHGSPQGPLAPPASPGPFATR
Inhibitor
Name:
BDBM557756
Synonyms:
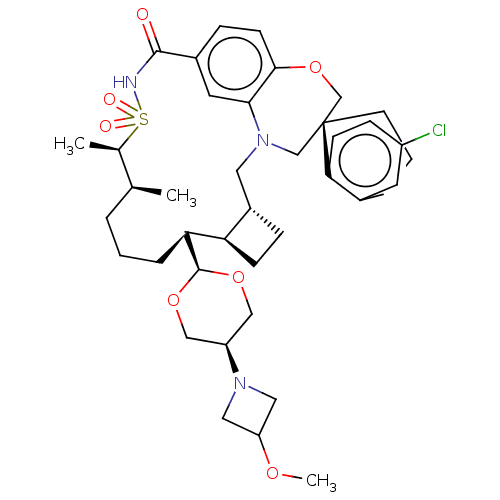
(1S,3'R,6'R,7'S,11'S,12'R)-6-chloro-7'-(cis-5- (3-methoxy-1-azetidinyl)-1,3-dioxan-2-yl)- 11',12'-dimethyl-3,4-dihydro-2H,15'H- spiro[naphthalene-1,22'- [20]oxa[13]thia[1,14]diazatetracyclo [14.7.2.0~3,6~.0~19,24~]pentacosa[16,18,24] trien]-15'-one 13',13'-dioxide OR | US11364248, Example 52 | US11364248, Example 53 | US11364248, Example 54
Type:
Small organic molecule
Emp. Form.:
C40H54ClN3O7S
Mol. Mass.:
756.391
SMILES:
COC1CN(C1)[C@H]1CO[C@H](OC1)[C@H]1CCC[C@H](C)[C@@H](C)S(=O)(=O)NC(=O)c2ccc3OC[C@]4(CCCc5cc(Cl)ccc45)CN(C[C@@H]4CC[C@@H]14)c3c2 |r,wU:49.52,46.50,12.13,16.18,18.20,9.9,6.6,wD:32.34,(-.79,9.36,;.55,8.59,;.55,7.05,;1.64,5.97,;.55,4.88,;-.54,5.97,;.55,3.34,;1.88,2.57,;1.88,1.03,;.55,.26,;-.79,1.03,;-.79,2.57,;.55,-1.28,;1.88,-2.05,;3.21,-1.28,;4.55,-2.05,;4.55,-3.59,;6.04,-3.2,;5.88,-4.36,;6.97,-3.27,;5.88,-5.9,;7.37,-5.51,;6.28,-7.39,;4.42,-6.86,;3.09,-6.09,;3.09,-4.55,;1.75,-6.86,;1.75,-8.4,;.42,-9.17,;-.92,-8.4,;-2.12,-9.36,;-3.62,-9.02,;-4.29,-7.63,;-5.06,-8.97,;-6.6,-8.97,;-7.37,-7.63,;-6.6,-6.3,;-7.37,-4.97,;-6.6,-3.63,;-7.37,-2.3,;-5.06,-3.63,;-4.29,-4.97,;-5.06,-6.3,;-3.62,-6.25,;-2.12,-5.9,;-2.12,-4.36,;-.79,-3.59,;.55,-4.36,;.55,-2.82,;-.79,-2.05,;-.92,-6.86,;.42,-6.09,)|