Reaction Details Report a problem with these data
Report a problem with these data
 Report a problem with these data
Report a problem with these dataTarget
Carbonic anhydrase 9
Ligand
BDBM11641
Substrate
BDBM10856
Meas. Tech.
CA Inhibition Assay
pH
7.5±n/a
Temperature
295.15±n/a K
Ki
14±n/a nM
Citation
 Menchise, V; De Simone, G; Alterio, V; Di Fiore, A; Pedone, C; Scozzafava, A; Supuran, CT Carbonic anhydrase inhibitors: stacking with Phe131 determines active site binding region of inhibitors as exemplified by the X-ray crystal structure of a membrane-impermeant antitumor sulfonamide complexed with isozyme II. J Med Chem 48:5721-7 (2005) [PubMed] Article
Menchise, V; De Simone, G; Alterio, V; Di Fiore, A; Pedone, C; Scozzafava, A; Supuran, CT Carbonic anhydrase inhibitors: stacking with Phe131 determines active site binding region of inhibitors as exemplified by the X-ray crystal structure of a membrane-impermeant antitumor sulfonamide complexed with isozyme II. J Med Chem 48:5721-7 (2005) [PubMed] Article Target
Name:
Carbonic anhydrase 9
Synonyms:
CA-IX | CA9 | CAH9_HUMAN | Carbonate dehydratase IX | Carbonic anhydrase 9 (CA IX) | Carbonic anhydrase 9 (CAIX) | Carbonic anhydrase 9 precursor | Carbonic anhydrase IX (CA IX) | Carbonic anhydrase IX (CAIX) | Carbonic anhydrases IX | Carbonic anhydrases; II & IX | G250 | MN | Membrane antigen MN | RCC-associated antigen G250
Type:
Enzyme
Mol. Mass.:
49669.03
Organism:
Homo sapiens (Human)
Description:
Catalytic domain of human cloned isozyme was used in the assay
Residue:
459
Sequence:
MAPLCPSPWLPLLIPAPAPGLTVQLLLSLLLLVPVHPQRLPRMQEDSPLGGGSSGEDDPLGEEDLPSEEDSPREEDPPGEEDLPGEEDLPGEEDLPEVKPKSEEEGSLKLEDLPTVEAPGDPQEPQNNAHRDKEGDDQSHWRYGGDPPWPRVSPACAGRFQSPVDIRPQLAAFCPALRPLELLGFQLPPLPELRLRNNGHSVQLTLPPGLEMALGPGREYRALQLHLHWGAAGRPGSEHTVEGHRFPAEIHVVHLSTAFARVDEALGRPGGLAVLAAFLEEGPEENSAYEQLLSRLEEIAEEGSETQVPGLDISALLPSDFSRYFQYEGSLTTPPCAQGVIWTVFNQTVMLSAKQLHTLSDTLWGPGDSRLQLNFRATQPLNGRVIEASFPAGVDSSPRAAEPVQLNSCLAAGDILALVFGLLFAVTSVAFLVQMRRQHRRGTKGGVSYRPAEVAETGA
Inhibitor
Name:
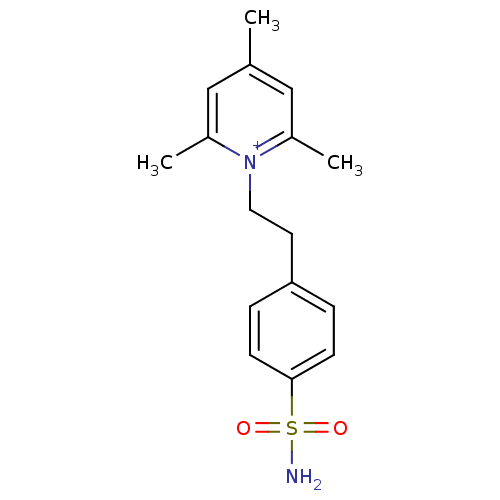
BDBM11641
Synonyms:
1-N-(4-sulfamoylphenyl-ethyl)-2,4,6-trimethylpyridiniumperchlorate | 2,4,6-trimethyl-1-[2-(4-sulfamoylphenyl)ethyl]pyridin-1-ium perchlorate | CHEMBL352535 | Compound 10 | pyridinium deriv. 10
Type:
Small organic molecule
Emp. Form.:
C16H21N2O2S
Mol. Mass.:
305.415
SMILES:
Cc1cc(C)[n+](CCc2ccc(cc2)S(N)(=O)=O)c(C)c1