Reaction Details Report a problem with these data
Report a problem with these data
 Report a problem with these data
Report a problem with these dataTarget
Serine protease 1
Ligand
BDBM12592
Substrate
BDBM12500
Meas. Tech.
Enzyme Assay and Determination of the Inhibition Constants.
pH
7.5±n/a
Temperature
295.15±n/a K
Ki
>2900±n/a nM
Citation
 Maignan, S; Guilloteau, JP; Choi-Sledeski, YM; Becker, MR; Ewing, WR; Pauls, HW; Spada, AP; Mikol, V Molecular structures of human factor Xa complexed with ketopiperazine inhibitors: preference for a neutral group in the S1 pocket. J Med Chem 46:685-90 (2003) [PubMed] Article
Maignan, S; Guilloteau, JP; Choi-Sledeski, YM; Becker, MR; Ewing, WR; Pauls, HW; Spada, AP; Mikol, V Molecular structures of human factor Xa complexed with ketopiperazine inhibitors: preference for a neutral group in the S1 pocket. J Med Chem 46:685-90 (2003) [PubMed] Article Target
Name:
Serine protease 1
Synonyms:
Beta-Trypsin | Cationic trypsin | PRSS1 | TRP1 | TRY1 | TRY1_BOVIN | TRYP1 | Trypsin | Trypsin I
Type:
Enzyme
Mol. Mass.:
25790.52
Organism:
Bos taurus (bovine)
Description:
P00760
Residue:
246
Sequence:
MKTFIFLALLGAAVAFPVDDDDKIVGGYTCGANTVPYQVSLNSGYHFCGGSLINSQWVVSAAHCYKSGIQVRLGEDNINVVEGNEQFISASKSIVHPSYNSNTLNNDIMLIKLKSAASLNSRVASISLPTSCASAGTQCLISGWGNTKSSGTSYPDVLKCLKAPILSDSSCKSAYPGQITSNMFCAGYLEGGKDSCQGDSGGPVVCSGKLQGIVSWGSGCAQKNKPGVYTKVCNYVSWIKQTIASN
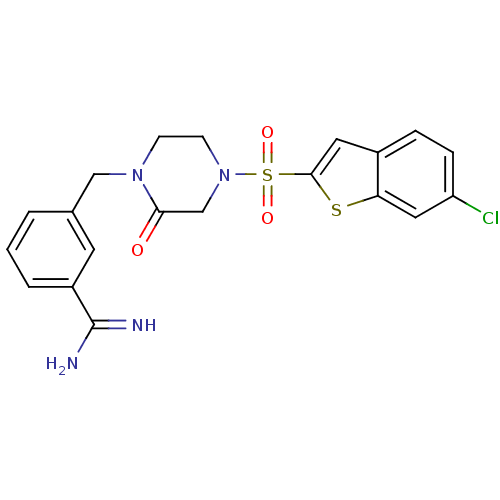
Inhibitor
Name:
BDBM12592
Synonyms:
3-({4-[(6-CHLORO-1-BENZOTHIEN-2-YL)SULFONYL]-2-OXOPIPERAZIN-1-YL}METHYL)BENZENECARBOXIMIDAMIDE | 3-({4-[(6-chloro-1-benzothiophene-2-)sulfonyl]-2-oxopiperazin-1-yl}methyl)benzene-1-carboximidamide | Ketopiperazine | RPR132747
Type:
Small organic molecule
Emp. Form.:
C20H19ClN4O3S2
Mol. Mass.:
462.973
SMILES:
NC(=N)c1cccc(CN2CCN(CC2=O)S(=O)(=O)c2cc3ccc(Cl)cc3s2)c1
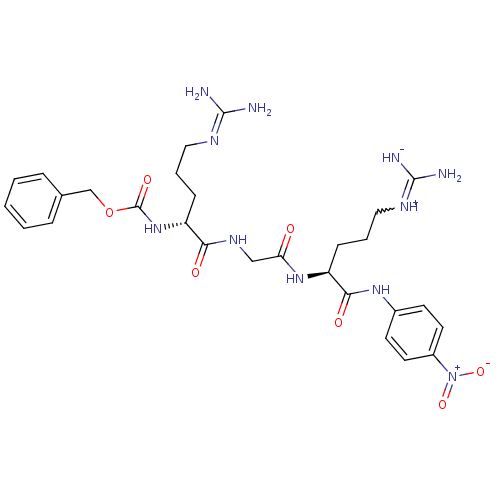
Substrate
Name:
BDBM12500
Synonyms:
Cbz-D-Arg-Gly-L-Arg-pNA | Chromogenic Substrate S-2765 | N-a-Benzyloxycarbonyl-Darginyl-L-glycyl-L-arginine-pnitroaniline | N-alpha-Z-D-Arg-Gly-Arg-p-nitroanilide | benzyl N-[(1R)-4-carbamimidamido-1-[({[(1S)-4-carbamimidamido-1-[(4-nitrophenyl)carbamoyl]butyl]carbamoyl}methyl)carbamoyl]butyl]carbamate
Type:
Small organic molecule
Emp. Form.:
C28H39N11O7
Mol. Mass.:
641.6788
SMILES:
[#7]\[#6](-[#7])=[#7]/[#6]-[#6]-[#6]-[#6@@H](-[#7]-[#6](=O)-[#8]-[#6]-c1ccccc1)-[#6](=O)-[#7]-[#6]-[#6](=O)-[#7]-[#6@@H](-[#6]-[#6]-[#6]-[#7+]=[#6](-[#7])-[#7-])-[#6](=O)-[#7]-c1ccc(cc1)-[#7+](-[#8-])=O |r,w:30.30|