Reaction Details Report a problem with these data
Report a problem with these data
 Report a problem with these data
Report a problem with these dataTarget
Serine protease 1
Ligand
BDBM13563
Substrate
BDBM13574
Meas. Tech.
Serine Protease Inhibition Assay
Ki
4000±n/a nM
Citation
 Zbinden, KG; Obst-Sander, U; Hilpert, K; Kuhne, H; Banner, DW; Bohm, HJ; Stahl, M; Ackermann, J; Alig, L; Weber, L; Wessel, HP; Riederer, MA; Tschopp, TB; Lave, T Selective and orally bioavailable phenylglycine tissue factor/factor VIIa inhibitors. Bioorg Med Chem Lett 15:5344-52 (2005) [PubMed] Article
Zbinden, KG; Obst-Sander, U; Hilpert, K; Kuhne, H; Banner, DW; Bohm, HJ; Stahl, M; Ackermann, J; Alig, L; Weber, L; Wessel, HP; Riederer, MA; Tschopp, TB; Lave, T Selective and orally bioavailable phenylglycine tissue factor/factor VIIa inhibitors. Bioorg Med Chem Lett 15:5344-52 (2005) [PubMed] Article More Info.:
Target
Name:
Serine protease 1
Synonyms:
Beta-Trypsin | Cationic trypsin | PRSS1 | TRP1 | TRY1 | TRY1_BOVIN | TRYP1 | Trypsin | Trypsin I
Type:
Enzyme
Mol. Mass.:
25790.52
Organism:
Bos taurus (bovine)
Description:
P00760
Residue:
246
Sequence:
MKTFIFLALLGAAVAFPVDDDDKIVGGYTCGANTVPYQVSLNSGYHFCGGSLINSQWVVSAAHCYKSGIQVRLGEDNINVVEGNEQFISASKSIVHPSYNSNTLNNDIMLIKLKSAASLNSRVASISLPTSCASAGTQCLISGWGNTKSSGTSYPDVLKCLKAPILSDSSCKSAYPGQITSNMFCAGYLEGGKDSCQGDSGGPVVCSGKLQGIVSWGSGCAQKNKPGVYTKVCNYVSWIKQTIASN
Inhibitor
Name:
BDBM13563
Synonyms:
2-[(4-carbamimidoylphenyl)amino]-2-{3-ethoxy-5-[(4-hydroxycyclohexyl)oxy]phenyl}acetic acid | phenylglycine deriv. 11
Type:
Small organic molecule
Emp. Form.:
C23H29N3O5
Mol. Mass.:
427.4935
SMILES:
CCOc1cc(O[C@H]2CC[C@H](O)CC2)cc(c1)C(Nc1ccc(cc1)C(N)=N)C(O)=O |r,wU:7.6,wD:10.10,(-7.94,9.1,;-6.61,8.33,;-5.27,9.1,;-3.94,8.33,;-2.61,9.1,;-1.27,8.33,;.06,9.1,;1.39,8.33,;2.73,9.1,;4.06,8.33,;4.06,6.79,;5.4,6.02,;2.73,6.02,;1.39,6.79,;-1.27,6.79,;-2.61,6.02,;-3.94,6.79,;-2.61,4.48,;-1.27,3.71,;-1.27,2.17,;-2.61,1.4,;-2.61,-.14,;-1.27,-.91,;.06,-.14,;.06,1.4,;-1.27,-2.45,;-2.61,-3.22,;.06,-3.22,;-3.94,3.71,;-5.27,4.48,;-3.94,2.17,)|
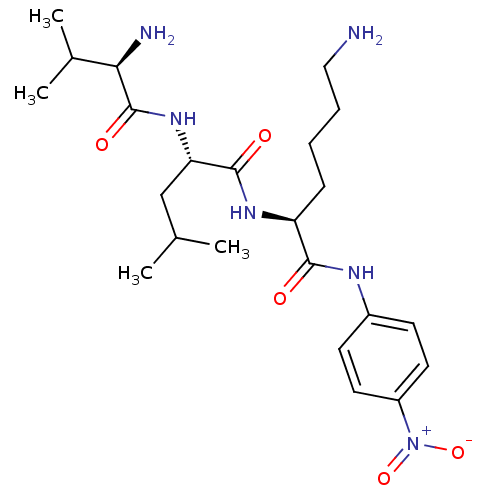
Substrate
Name:
BDBM13574
Synonyms:
(2S)-6-amino-2-[(2S)-2-[(2R)-2-amino-3-methylbutanamido]-4-methylpentanamido]-N-(4-nitrophenyl)hexanamide dihydrochloride | Chromogenic Substrate S-2251 | H-D-Valyl-L-leucyl-L-lysine-p-Nitroaniline dihydrochloride
Type:
Small organic molecule
Emp. Form.:
C23H38N6O5
Mol. Mass.:
478.585
SMILES:
CC(C)C[C@H](NC(=O)[C@H](N)C(C)C)C(=O)N[C@@H](CCCCN)C(=O)Nc1ccc(cc1)[N+]([O-])=O |r|