Reaction Details Report a problem with these data
Report a problem with these data
 Report a problem with these data
Report a problem with these dataTarget
Dimer of Gag-Pol polyprotein [600-1159,L699I,K702N,C879S]
Ligand
BDBM16196
Substrate
poly(rA).oligo(dT)12-18
Meas. Tech.
HIV-1 RT Assay
pH
7.5±n/a
Temperature
310.15±n/a K
IC50
19800±5400 nM
Citation
 Herschhorn, A; Lerman, L; Weitman, M; Gleenberg, IO; Nudelman, A; Hizi, A De Novo Parallel Design, Synthesis and Evaluation of Inhibitors against the Reverse Transcriptase of Human Immunodeficiency Virus Type-1 and Drug-Resi J Med Chem 50:2370-84 (2007) [PubMed] Article
Herschhorn, A; Lerman, L; Weitman, M; Gleenberg, IO; Nudelman, A; Hizi, A De Novo Parallel Design, Synthesis and Evaluation of Inhibitors against the Reverse Transcriptase of Human Immunodeficiency Virus Type-1 and Drug-Resi J Med Chem 50:2370-84 (2007) [PubMed] Article Target
Name:
Dimer of Gag-Pol polyprotein [600-1159,L699I,K702N,C879S]
Synonyms:
HIV-1 Reverse Transcriptase Mutant (L100I/K103N)
Type:
Protein Complex
Mol. Mass.:
n/a
Description:
The double mutant L100I/K103N was expressed in E. coli as a homodimer enzyme with two identical 66 kDa subunits.
Components:
This complex has 2 components.
Component 1
Name:
Gag-Pol polyprotein [600-1159,L699I,K702N,C879S]
Synonyms:
HIV-1 Reverse Transcriptase Mutant (L100I/K103N) Chain A | HIV-1 Reverse Transcriptase Mutant (L100I/K103N) Chain B | POL_HV1B1 | gag-pol
Type:
Enzyme Subunit
Mol. Mass.:
64410.35
Organism:
Human immunodeficiency virus type 1
Description:
P03366[600-1159,L699I,K702N,C879S]
Residue:
560
Sequence:
PISPIETVPVKLKPGMDGPKVKQWPLTEEKIKALVEICTEMEKEGKISKIGPENPYNTPVFAIKKKDSTKWRKLVDFRELNKRTQDFWEVQLGIPHPAGIKKNKSVTVLDVGDAYFSVPLDEDFRKYTAFTIPSINNETPGIRYQYNVLPQGWKGSPAIFQSSMTKILEPFKKQNPDIVIYQYMDDLYVGSDLEIGQHRTKIEELRQHLLRWGLTTPDKKHQKEPPFLWMGYELHPDKWTVQPIVLPEKDSWTVNDIQKLVGKLNWASQIYPGIKVRQLSKLLRGTKALTEVIPLTEEAELELAENREILKEPVHGVYYDPSKDLIAEIQKQGQGQWTYQIYQEPFKNLKTGKYARMRGAHTNDVKQLTEAVQKITTESIVIWGKTPKFKLPIQKETWETWWTEYWQATWIPEWEFVNTPPLVKLWYQLEKEPIVGAETFYVDGAANRETKLGKAGYVTNKGRQKVVPLTNTTNQKTELQAIYLALQDSGLEVNIVTDSQYALGIIQAQPDKSESELVNQIIEQLIKKEKVYLAWVPAHKGIGGNEQVDKLVSAGIRKIL
Component 2
Name:
Gag-Pol polyprotein [600-1159,L699I,K702N,C879S]
Synonyms:
HIV-1 Reverse Transcriptase Mutant (L100I/K103N) Chain A | HIV-1 Reverse Transcriptase Mutant (L100I/K103N) Chain B | POL_HV1B1 | gag-pol
Type:
Enzyme Subunit
Mol. Mass.:
64410.35
Organism:
Human immunodeficiency virus type 1
Description:
P03366[600-1159,L699I,K702N,C879S]
Residue:
560
Sequence:
PISPIETVPVKLKPGMDGPKVKQWPLTEEKIKALVEICTEMEKEGKISKIGPENPYNTPVFAIKKKDSTKWRKLVDFRELNKRTQDFWEVQLGIPHPAGIKKNKSVTVLDVGDAYFSVPLDEDFRKYTAFTIPSINNETPGIRYQYNVLPQGWKGSPAIFQSSMTKILEPFKKQNPDIVIYQYMDDLYVGSDLEIGQHRTKIEELRQHLLRWGLTTPDKKHQKEPPFLWMGYELHPDKWTVQPIVLPEKDSWTVNDIQKLVGKLNWASQIYPGIKVRQLSKLLRGTKALTEVIPLTEEAELELAENREILKEPVHGVYYDPSKDLIAEIQKQGQGQWTYQIYQEPFKNLKTGKYARMRGAHTNDVKQLTEAVQKITTESIVIWGKTPKFKLPIQKETWETWWTEYWQATWIPEWEFVNTPPLVKLWYQLEKEPIVGAETFYVDGAANRETKLGKAGYVTNKGRQKVVPLTNTTNQKTELQAIYLALQDSGLEVNIVTDSQYALGIIQAQPDKSESELVNQIIEQLIKKEKVYLAWVPAHKGIGGNEQVDKLVSAGIRKIL
Inhibitor
Name:
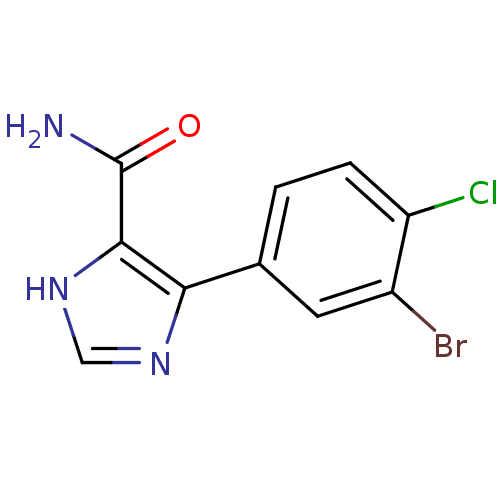
BDBM16196
Synonyms:
4-(3-Bromo-4-chlorophenyl)-1H-imidazole-5-carboxamide | Imidazole 8c
Type:
Small organic molecule
Emp. Form.:
C10H7BrClN3O
Mol. Mass.:
300.539
SMILES:
NC(=O)c1[nH]cnc1-c1ccc(Cl)c(Br)c1
Substrate
Name:
poly(rA).oligo(dT)12-18
Synonyms:
n/a
Type:
RNA/DNA duplex
Mol. Mass.:
358.43
Organism:
n/a
Description:
The incorporation of [3H]dTTP into poly(rA).oligo(dT)12-18 template-primer.
Residue:
3
Sequence:
NA