Reaction Details Report a problem with these data
Report a problem with these data
 Report a problem with these data
Report a problem with these dataTarget
Peptide deformylase
Ligand
BDBM21684
Substrate
PDF substrate peptide
Meas. Tech.
PDF Inhibition Assay
pH
7.6±n/a
Temperature
298.15±n/a K
IC50
1150±80 nM
Km
9100000±n/a nM
kcat
103±n/a 1/sec
Citation
 Smith, KJ; Petit, CM; Aubart, K; Smyth, M; McManus, E; Jones, J; Fosberry, A; Lewis, C; Lonetto, M; Christensen, SB Structural variation and inhibitor binding in polypeptide deformylase from four different bacterial species. Protein Sci 12:349-60 (2003) [PubMed] Article
Smith, KJ; Petit, CM; Aubart, K; Smyth, M; McManus, E; Jones, J; Fosberry, A; Lewis, C; Lonetto, M; Christensen, SB Structural variation and inhibitor binding in polypeptide deformylase from four different bacterial species. Protein Sci 12:349-60 (2003) [PubMed] Article Target
Name:
Peptide deformylase
Synonyms:
DEF_ECOLI | PDF | Peptide Deformylase | Polypeptide deformylase | def | fms
Type:
Enzyme
Mol. Mass.:
19323.16
Organism:
Escherichia coli (strain K12)
Description:
The pdf gene from E. coli K12 was cloned and protein was expressed and purified from E. coli BL21(DE3).
Residue:
169
Sequence:
MSVLQVLHIPDERLRKVAKPVEEVNAEIQRIVDDMFETMYAEEGIGLAATQVDIHQRIIVIDVSENRDERLVLINPELLEKSGETGIEEGCLSIPEQRALVPRAEKVKIRALDRDGKPFELEADGLLAICIQHEMDHLVGKLFMDYLSPLKQQRIRQKVEKLDRLKARA
Inhibitor
Name:
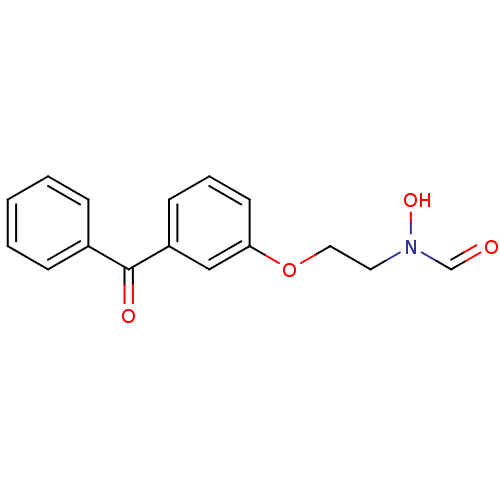
BDBM21684
Synonyms:
N-[2-(3-benzoylphenoxy)ethyl]-N-hydroxyformamide | SB-543668
Type:
Small organic molecule
Emp. Form.:
C16H15NO4
Mol. Mass.:
285.2946
SMILES:
ON(CCOc1cccc(c1)C(=O)c1ccccc1)C=O
Substrate
Name:
PDF substrate peptide
Synonyms:
n/a
Type:
Peptide
Mol. Mass.:
1718.01
Organism:
n/a
Description:
n/a
Residue:
14
Sequence:
FRMYLMETALASER