Reaction Details Report a problem with these data
Report a problem with these data
 Report a problem with these data
Report a problem with these dataTarget
COUP transcription factor 1
Ligand
BDBM106870
Substrate
n/a
Meas. Tech.
ChEMBL_1544013 (CHEMBL3750385)
IC50
>10000±n/a nM
Citation
 Burger, MT; Nishiguchi, G; Han, W; Lan, J; Simmons, R; Atallah, G; Ding, Y; Tamez, V; Zhang, Y; Mathur, M; Muller, K; Bellamacina, C; Lindvall, MK; Zang, R; Huh, K; Feucht, P; Zavorotinskaya, T; Dai, Y; Basham, S; Chan, J; Ginn, E; Aycinena, A; Holash, J; Castillo, J; Langowski, JL; Wang, Y; Chen, MY; Lambert, A; Fritsch, C; Kauffmann, A; Pfister, E; Vanasse, KG; Garcia, PD Identification of N-(4-((1R,3S,5S)-3-Amino-5-methylcyclohexyl)pyridin-3-yl)-6-(2,6-difluorophenyl)-5-fluoropicolinamide (PIM447), a Potent and Selective Proviral Insertion Site of Moloney Murine Leukemia (PIM) 1, 2, and 3 Kinase Inhibitor in Clinical Trials for Hematological Malignancies. J Med Chem 58:8373-86 (2015) [PubMed] Article
Burger, MT; Nishiguchi, G; Han, W; Lan, J; Simmons, R; Atallah, G; Ding, Y; Tamez, V; Zhang, Y; Mathur, M; Muller, K; Bellamacina, C; Lindvall, MK; Zang, R; Huh, K; Feucht, P; Zavorotinskaya, T; Dai, Y; Basham, S; Chan, J; Ginn, E; Aycinena, A; Holash, J; Castillo, J; Langowski, JL; Wang, Y; Chen, MY; Lambert, A; Fritsch, C; Kauffmann, A; Pfister, E; Vanasse, KG; Garcia, PD Identification of N-(4-((1R,3S,5S)-3-Amino-5-methylcyclohexyl)pyridin-3-yl)-6-(2,6-difluorophenyl)-5-fluoropicolinamide (PIM447), a Potent and Selective Proviral Insertion Site of Moloney Murine Leukemia (PIM) 1, 2, and 3 Kinase Inhibitor in Clinical Trials for Hematological Malignancies. J Med Chem 58:8373-86 (2015) [PubMed] Article More Info.:
Target
Name:
COUP transcription factor 1
Synonyms:
COT1_HUMAN | COUP transcription factor I | COUP-TF I | COUP-TF1 | EAR-3 | EAR3 | ERBAL3 | NR2F1 | Nuclear receptor subfamily 2 group F member 1 | TFCOUP1 | V-erbA-related protein 3
Type:
PROTEIN
Mol. Mass.:
46167.74
Organism:
Homo sapiens (Human)
Description:
ChEMBL_876574
Residue:
423
Sequence:
MAMVVSSWRDPQDDVAGGNPGGPNPAAQAARGGGGGAGEQQQQAGSGAPHTPQTPGQPGAPATPGTAGDKGQGPPGSGQSQQHIECVVCGDKSSGKHYGQFTCEGCKSFFKRSVRRNLTYTCRANRNCPIDQHHRNQCQYCRLKKCLKVGMRREAVQRGRMPPTQPNPGQYALTNGDPLNGHCYLSGYISLLLRAEPYPTSRYGSQCMQPNNIMGIENICELAARLLFSAVEWARNIPFFPDLQITDQVSLLRLTWSELFVLNAAQCSMPLHVAPLLAAAGLHASPMSADRVVAFMDHIRIFQEQVEKLKALHVDSAEYSCLKAIVLFTSDACGLSDAAHIESLQEKSQCALEEYVRSQYPNQPSRFGKLLLRLPSLRTVSSSVIEQLFFVRLVGKTPIETLIRDMLLSGSSFNWPYMSIQCS
Inhibitor
Name:
BDBM106870
Synonyms:
US8592455, 70
Type:
Small organic molecule
Emp. Form.:
C24H23F3N4O
Mol. Mass.:
440.4608
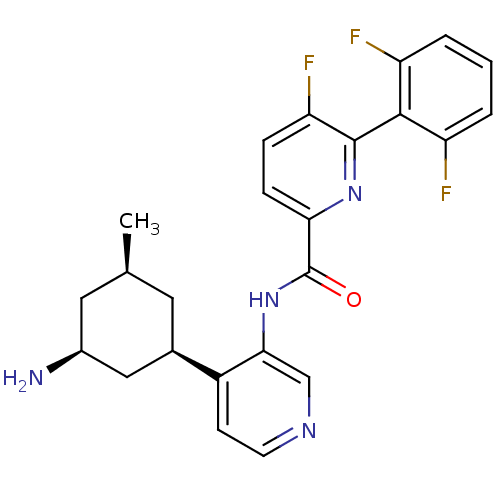
SMILES:
C[C@@H]1C[C@H](N)C[C@@H](C1)c1ccncc1NC(=O)c1ccc(F)c(n1)-c1c(F)cccc1F |r,wU:6.8,3.3,1.0,(-.67,2.69,;-2,1.93,;-3.33,2.69,;-4.67,1.93,;-6,2.69,;-4.67,.38,;-3.33,-.38,;-2,.38,;-3.33,-1.93,;-4.67,-2.69,;-4.67,-4.23,;-3.33,-5,;-2,-4.23,;-2,-2.69,;-.67,-1.93,;.67,-2.69,;.67,-4.23,;2,-1.93,;3.33,-2.69,;4.67,-1.93,;4.67,-.38,;6,.38,;3.33,.38,;2,-.38,;3.33,1.93,;4.67,2.69,;6,1.93,;4.67,4.23,;3.33,5,;2,4.23,;2,2.69,;.67,1.93,)|