Reaction Details Report a problem with these data
Report a problem with these data
 Report a problem with these data
Report a problem with these dataTarget
Fatty-acid amide hydrolase 1 [30-579]
Ligand
BDBM143359
Substrate
n/a
Meas. Tech.
ChEMBL_1870781 (CHEMBL4371948)
IC50
12±n/a nM
Citation
 Bhuniya, D; Kharul, RK; Hajare, A; Shaikh, N; Bhosale, S; Balwe, S; Begum, F; De, S; Athavankar, S; Joshi, D; Madgula, V; Joshi, K; Raje, AA; Meru, AV; Magdum, A; Mookhtiar, KA; Barbhaiya, R Discovery and evaluation of novel FAAH inhibitors in neuropathic pain model. Bioorg Med Chem Lett 29:238-243 (2019) [PubMed] Article
Bhuniya, D; Kharul, RK; Hajare, A; Shaikh, N; Bhosale, S; Balwe, S; Begum, F; De, S; Athavankar, S; Joshi, D; Madgula, V; Joshi, K; Raje, AA; Meru, AV; Magdum, A; Mookhtiar, KA; Barbhaiya, R Discovery and evaluation of novel FAAH inhibitors in neuropathic pain model. Bioorg Med Chem Lett 29:238-243 (2019) [PubMed] Article More Info.:
Target
Name:
Fatty-acid amide hydrolase 1 [30-579]
Synonyms:
Anandamide amidohydrolase 1 | FAAH1_RAT | Faah | Faah1 | Fatty Acid Amide Hydrolase | Fatty Acid Amide Hydrolic, FAAH | Fatty-acid amide hydrolase (FAAH) | Fatty-acid amide hydrolase 1 | Fatty-acid amide hydrolase 1 (FAAH) | Fatty-acid amide hydrolase 1 (aa 30-579) | Oleamide hydrolase 1
Type:
Single-pass membrane protein; homodimer
Mol. Mass.:
60474.00
Organism:
Rattus norvegicus (rat)
Description:
P97612 (aa 30-579)
Residue:
550
Sequence:
RWTGRQKARGAATRARQKQRASLETMDKAVQRFRLQNPDLDSEALLTLPLLQLVQKLQSGELSPEAVFFTYLGKAWEVNKGTNCVTSYLTDCETQLSQAPRQGLLYGVPVSLKECFSYKGHDSTLGLSLNEGMPSESDCVVVQVLKLQGAVPFVHTNVPQSMLSFDCSNPLFGQTMNPWKSSKSPGGSSGGEGALIGSGGSPLGLGTDIGGSIRFPSAFCGICGLKPTGNRLSKSGLKGCVYGQTAVQLSLGPMARDVESLALCLKALLCEHLFTLDPTVPPLPFREEVYRSSRPLRVGYYETDNYTMPSPAMRRALIETKQRLEAAGHTLIPFLPNNIPYALEVLSAGGLFSDGGRSFLQNFKGDFVDPCLGDLILILRLPSWFKRLLSLLLKPLFPRLAAFLNSMRPRSAEKLWKLQHEIEMYRQSVIAQWKAMNLDVLLTPMLGPALDLNTPGRATGAISYTVLYNCLDFPAGVVPVTTVTAEDDAQMELYKGYFGDIWDIILKKAMKNSVGLPVAVQCVALPWQEELCLRFMREVEQLMTPQKQPS
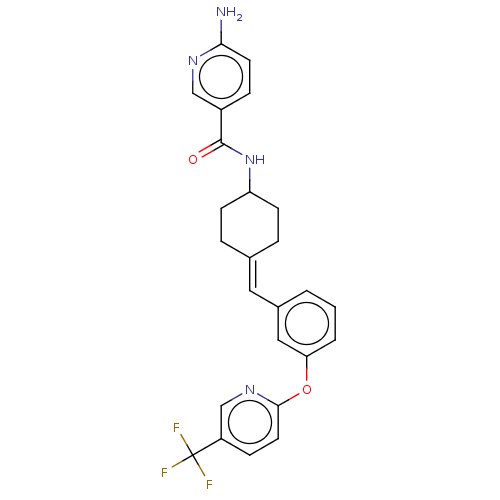
Inhibitor
Name:
BDBM143359
Synonyms:
US9682953, 20.A-3
Type:
Small organic molecule
Emp. Form.:
C25H23F3N4O2
Mol. Mass.:
468.4709
SMILES:
Nc1ccc(cn1)C(=O)NC1CCC(CC1)=Cc1cccc(Oc2ccc(cn2)C(F)(F)F)c1 |(12.65,-1.17,;11.31,-1.94,;11.31,-3.48,;9.98,-4.25,;8.65,-3.48,;8.65,-1.94,;9.98,-1.17,;7.31,-4.25,;7.31,-5.79,;5.98,-3.48,;4.65,-4.25,;4.65,-5.79,;3.31,-6.56,;1.98,-5.79,;1.98,-4.25,;3.31,-3.48,;.65,-6.56,;-.69,-5.79,;-.69,-4.25,;-2.02,-3.48,;-3.36,-4.25,;-3.36,-5.79,;-4.69,-6.56,;-6.02,-5.79,;-6.02,-4.25,;-7.36,-3.48,;-8.69,-4.25,;-8.69,-5.79,;-7.36,-6.56,;-10.02,-3.48,;-11.36,-4.25,;-10.02,-1.94,;-10.02,-5.02,;-2.02,-6.56,)|