Reaction Details Report a problem with these data
Report a problem with these data
 Report a problem with these data
Report a problem with these dataTarget
Peptide deformylase
Ligand
BDBM50099857
Substrate
n/a
Meas. Tech.
ChEBML_154331
IC50
>100000±n/a nM
Citation
 Thorarensen, A; Deibel, MR; Rohrer, DC; Vosters, AF; Yem, AW; Marshall, VD; Lynn, JC; Bohanon, MJ; Tomich, PK; Zurenko, GE; Sweeney, MT; Jensen, RM; Nielsen, JW; Seest, EP; Dolak, LA Identification of novel potent hydroxamic acid inhibitors of peptidyl deformylase and the importance of the hydroxamic acid functionality on inhibition. Bioorg Med Chem Lett 11:1355-8 (2001) [PubMed] Article
Thorarensen, A; Deibel, MR; Rohrer, DC; Vosters, AF; Yem, AW; Marshall, VD; Lynn, JC; Bohanon, MJ; Tomich, PK; Zurenko, GE; Sweeney, MT; Jensen, RM; Nielsen, JW; Seest, EP; Dolak, LA Identification of novel potent hydroxamic acid inhibitors of peptidyl deformylase and the importance of the hydroxamic acid functionality on inhibition. Bioorg Med Chem Lett 11:1355-8 (2001) [PubMed] Article More Info.:
Target
Name:
Peptide deformylase
Synonyms:
DEF_STAAU | PDF | Peptide Deformylase | Polypeptide deformylase | def | def1 | pdf1
Type:
PROTEIN
Mol. Mass.:
20556.80
Organism:
Staphylococcus aureus (strain Mu50 / ATCC 700699)
Description:
ChEMBL_459563
Residue:
183
Sequence:
MLTMKDIIRDGHPTLRQKAAELELPLTKEEKETLIAMREFLVNSQDEEIAKRYGLRSGVGLAAPQINISKRMIAVLIPDDGSGKSYDYMLVNPKIVSHSVQEAYLPTGEGCLSVDDNVAGLVHRHNRITIKAKDIEGNDIQLRLKGYPAIVFQHEIDHLNGVMFYDHIDKNHPLQPHTDAVEV
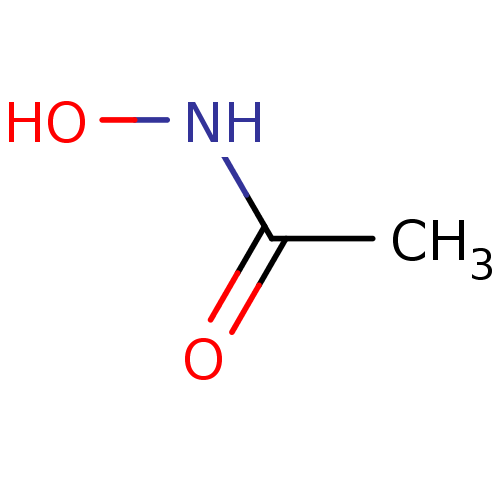
Inhibitor
Name:
BDBM50099857
Synonyms:
ACETOHYDROXAMIC ACID (AHA) | AHA | Acethydroxamsaeure | Acethydroxamsaure | Acetic acid, oxime | Acetylhydroxamic acid | Cetohyroxamic acid | Lithostat | Methylhydroxamic acid | N-Acetyl hydroxyacetamide | N-Acetylhydroxylamine | N-Hydroxyacetamide | acetohydroxamic acid
Type:
Small organic molecule
Emp. Form.:
C2H5NO2
Mol. Mass.:
75.0666
SMILES:
CC(=O)NO