Reaction Details Report a problem with these data
Report a problem with these data
 Report a problem with these data
Report a problem with these dataTarget
Prosaposin
Ligand
BDBM50092055
Substrate
n/a
Meas. Tech.
ChEMBL_2045826 (CHEMBL4700525)
Kd
6900±n/a nM
Citation
 Milliken, BT; Melegari, L; Smith, GL; Grohn, K; Wolfe, AJ; Moody, K; Bou-Abdallah, F; Doyle, RP Fenretinide binding to the lysosomal protein saposin D alters ceramide solubilization and hydrolysis. RSC Med Chem 11:1048-1052 (2020) [PubMed] Article
Milliken, BT; Melegari, L; Smith, GL; Grohn, K; Wolfe, AJ; Moody, K; Bou-Abdallah, F; Doyle, RP Fenretinide binding to the lysosomal protein saposin D alters ceramide solubilization and hydrolysis. RSC Med Chem 11:1048-1052 (2020) [PubMed] Article More Info.:
Target
Name:
Prosaposin
Synonyms:
A1 activator | CSAct | Cerebroside sulfate activator | Co-beta-glucosidase | Component C | Dispersin | GLBA | Glucosylceramidase activator | PSAP | Proactivator polypeptide | Prosaposin | Protein A | Protein C | SAP-1 | SAP-2 | SAP1 | SAP_HUMAN | Saposin-A | Saposin-B | Saposin-B-Val | Saposin-C | Saposin-D | Sphingolipid activator protein 1 | Sphingolipid activator protein 2 | Sulfatide/GM1 activator
Type:
PROTEIN
Mol. Mass.:
58096.49
Organism:
Homo sapiens
Description:
ChEMBL_109808
Residue:
524
Sequence:
MYALFLLASLLGAALAGPVLGLKECTRGSAVWCQNVKTASDCGAVKHCLQTVWNKPTVKSLPCDICKDVVTAAGDMLKDNATEEEILVYLEKTCDWLPKPNMSASCKEIVDSYLPVILDIIKGEMSRPGEVCSALNLCESLQKHLAELNHQKQLESNKIPELDMTEVVAPFMANIPLLLYPQDGPRSKPQPKDNGDVCQDCIQMVTDIQTAVRTNSTFVQALVEHVKEECDRLGPGMADICKNYISQYSEIAIQMMMHMQPKEICALVGFCDEVKEMPMQTLVPAKVASKNVIPALELVEPIKKHEVPAKSDVYCEVCEFLVKEVTKLIDNNKTEKEILDAFDKMCSKLPKSLSEECQEVVDTYGSSILSILLEEVSPELVCSMLHLCSGTRLPALTVHVTQPKDGGFCEVCKKLVGYLDRNLEKNSTKQEILAALEKGCSFLPDPYQKQCDQFVAEYEPVLIEILVEVMDPSFVCLKIGACPSAHKPLLGTEKCIWGPSYWCQNTETAAQCNAVEHCKRHVWN
Inhibitor
Name:
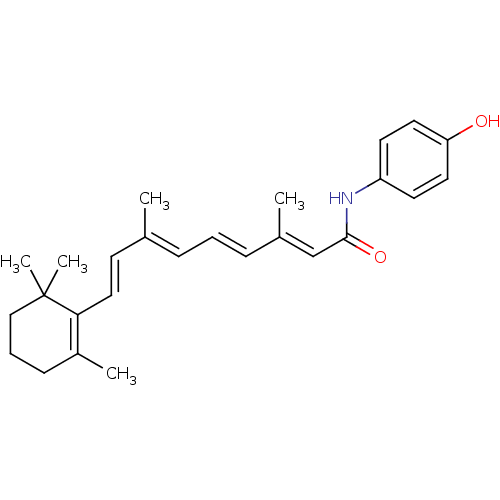
BDBM50092055
Synonyms:
(2E,4E,6E,8E)-3,7-Dimethyl-9-(2,6,6-trimethyl-cyclohex-1-enyl)-nona-2,4,6,8-tetraenoic acid (4-hydroxy-phenyl)-amide | (2E,4E,6E,8E)-N-(4-hydroxyphenyl)-3,7-dimethyl-9-(2,6,6-trimethylcyclohex-1-enyl)nona-2,4,6,8-tetraenamide | 3,7-Dimethyl-9-(2,6,6-trimethyl-cyclohex-1-enyl)-nona-2,4,6,8-tetraenoic acid (4-hydroxy-phenyl)-amide | 4-hydroxyphenyl retinamide | CHEMBL7301 | FENRETINIDE | N-(4-HYDROXYPHENYL)ALL-TRANS RETINAMIDE | N-(4-hydroxyphenyl)-retinamide | N-(4-hydroxyphenyl)retinamide | N-(4-hydroxyphenyl)retinamide, 4-HPR
Type:
Small organic molecule
Emp. Form.:
C26H33NO2
Mol. Mass.:
391.5457
SMILES:
C\C(\C=C\C1=C(C)CCCC1(C)C)=C/C=C/C(/C)=C/C(=O)Nc1ccc(O)cc1 |c:4|