Reaction Details Report a problem with these data
Report a problem with these data
 Report a problem with these data
Report a problem with these dataTarget
Genome polyprotein/Non-structural protein 4A
Ligand
BDBM50120642
Substrate
n/a
Meas. Tech.
ChEMBL_143646 (CHEMBL752805)
IC50
7000±n/a nM
Citation
 Nizi, E; Koch, U; Ponzi, S; Matassa, VG; Gardelli, C Capped dipeptide alpha-ketoacid inhibitors of the HCV NS3 protease. Bioorg Med Chem Lett 12:3325-8 (2002) [PubMed] Article
Nizi, E; Koch, U; Ponzi, S; Matassa, VG; Gardelli, C Capped dipeptide alpha-ketoacid inhibitors of the HCV NS3 protease. Bioorg Med Chem Lett 12:3325-8 (2002) [PubMed] Article More Info.:
Target
Name:
Genome polyprotein/Non-structural protein 4A
Synonyms:
Hepatitis C virus serine protease, NS3/NS4A
Type:
n/a
Mol. Mass.:
n/a
Description:
ASSAY_ID of ChEMBL is 1969955
Components:
This complex has 2 components.
Component 1
Name:
Non-structural protein 4A
Synonyms:
Hepatitis C virus NS4A protein | Hepatitis C virus serine protease, NS3/NS4A | Non-structural protein 4A
Type:
PROTEIN
Mol. Mass.:
5762.65
Organism:
Hepatitis C virus
Description:
ChEMBL_305334
Residue:
54
Sequence:
STWVLLGGVLAALAAYCLSVGCVVIVGYIELGGKPALVPDKEVCYQQYDEMEEC
Component 2
Name:
Genome polyprotein
Synonyms:
Hepatitis C virus NS3 protease/helicase | Hepatitis C virus serine protease, NS3/NS4A
Type:
Protein
Mol. Mass.:
67067.41
Organism:
Hepatitis C virus
Description:
A3EZI9
Residue:
631
Sequence:
APITAYAQQTRGLLGCIITSLTGRDKNQVEGEVQIVSTAAQTFLATCINGVCWTVYHGAGTRTIASSKGPVIQMYTNVDQDLVGWPAPQGARSLTPCTCGSSDLYLVTRHADVIPVRRRGDGRGSLLSPRPISYLKGSSGGPLLCPAGHAVGIFRAAVCTRGVAKAVDFIPVEGLETTMRSPVFSDNSSPPAVPQSYQVAHLHAPTGSGKSTKVPAAYAAQGYKVLVLNPSVAATLGFGAYMSKAHGIDPNIRTGVRTITTGSPITYSTYGKFLADGGCSGGAYDIIICDECHSTDATSILGIGTVLDQAETAGARLTVLATATPPGSVTVPHPNIEEVALSTTGEIPFYGKAIPLEAIKGGRHLIFCHSKKKCDELAAKLVALGVNAVAYYRGLDVSVIPASGDVVVVATDALMTGFTGDFDSVIDCNTCVTQTVDFSLDPTFTIETTTLPQDAVSRTQRRGRTGRGKPGIYRFVTPGERPSGMFDSSVLCECYDAGCAWYELTPAETTVRLRAYMNTPGLPVCQDHLEFWEGVFTGLTHIDAHFLSQTKQSGENLPYLVAYQATVCARAQAPPPSWDQMWKCLIRLKPTLHGPTPLLYRLGAVQNEITLTHPITKYIMTCMSADLEVVT
Inhibitor
Name:
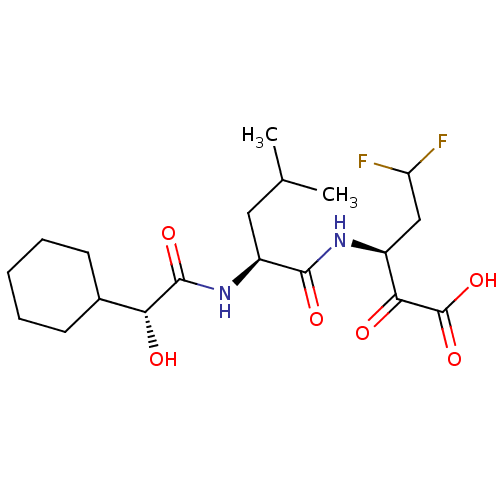
BDBM50120642
Synonyms:
3-[2-((R)-2-Cyclohexyl-2-hydroxy-1-(S)-oxo-ethylamino)-4-methyl-pentanoylamino]-5,5-difluoro-2-(S)-oxo-pentanoic acid | CHEMBL420303
Type:
Small organic molecule
Emp. Form.:
C19H30F2N2O6
Mol. Mass.:
420.4481
SMILES:
CC(C)C[C@H](NC(=O)[C@H](O)C1CCCCC1)C(=O)N[C@@H](CC(F)F)C(=O)C(O)=O