Reaction Details Report a problem with these data
Report a problem with these data
 Report a problem with these data
Report a problem with these dataTarget
MAP kinase-interacting serine/threonine-protein kinase 2
Ligand
BDBM5446
Substrate
n/a
Meas. Tech.
ChEMBL_325017 (CHEMBL860651)
Kd
1600±n/a nM
Citation
 Fabian, MA; Biggs, WH; Treiber, DK; Atteridge, CE; Azimioara, MD; Benedetti, MG; Carter, TA; Ciceri, P; Edeen, PT; Floyd, M; Ford, JM; Galvin, M; Gerlach, JL; Grotzfeld, RM; Herrgard, S; Insko, DE; Insko, MA; Lai, AG; Lélias, JM; Mehta, SA; Milanov, ZV; Velasco, AM; Wodicka, LM; Patel, HK; Zarrinkar, PP; Lockhart, DJ A small molecule-kinase interaction map for clinical kinase inhibitors. Nat Biotechnol 23:329-36 (2005) [PubMed] Article
Fabian, MA; Biggs, WH; Treiber, DK; Atteridge, CE; Azimioara, MD; Benedetti, MG; Carter, TA; Ciceri, P; Edeen, PT; Floyd, M; Ford, JM; Galvin, M; Gerlach, JL; Grotzfeld, RM; Herrgard, S; Insko, DE; Insko, MA; Lai, AG; Lélias, JM; Mehta, SA; Milanov, ZV; Velasco, AM; Wodicka, LM; Patel, HK; Zarrinkar, PP; Lockhart, DJ A small molecule-kinase interaction map for clinical kinase inhibitors. Nat Biotechnol 23:329-36 (2005) [PubMed] Article More Info.:
Target
Name:
MAP kinase-interacting serine/threonine-protein kinase 2
Synonyms:
GPRK7 | MAP kinase signal-integrating kinase 2 | MAP kinase-interacting serine/threonine-protein kinase 2 (MKNK2) | MAP kinase-interacting serine/threonine-protein kinase 2 (MnK2) | MAP-kinase interacting kinase 2 (MNK2) | MKNK2 | MKNK2_HUMAN | MNK2
Type:
Protein
Mol. Mass.:
51870.79
Organism:
Homo sapiens (Human)
Description:
Q9HBH9
Residue:
465
Sequence:
MVQKKPAELQGFHRSFKGQNPFELAFSLDQPDHGDSDFGLQCSARPDMPASQPIDIPDAKKRGKKKKRGRATDSFSGRFEDVYQLQEDVLGEGAHARVQTCINLITSQEYAVKIIEKQPGHIRSRVFREVEMLYQCQGHRNVLELIEFFEEEDRFYLVFEKMRGGSILSHIHKRRHFNELEASVVVQDVASALDFLHNKGIAHRDLKPENILCEHPNQVSPVKICDFDLGSGIKLNGDCSPISTPELLTPCGSAEYMAPEVVEAFSEEASIYDKRCDLWSLGVILYILLSGYPPFVGRCGSDCGWDRGEACPACQNMLFESIQEGKYEFPDKDWAHISCAAKDLISKLLVRDAKQRLSAAQVLQHPWVQGCAPENTLPTPMVLQRNSCAKDLTSFAAEAIAMNRQLAQHDEDLAEEEAAGQGQPVLVRATSRCLQLSPPSQSKLAQRRQRASLSSAPVVLVGDHA
Inhibitor
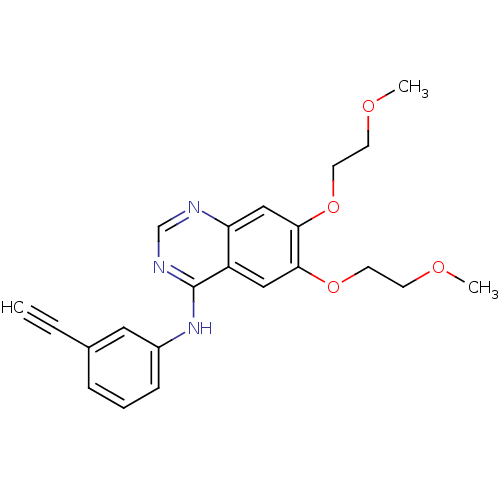
Name:
BDBM5446
Synonyms:
CHEMBL553 | ERLOTINIB HYDROCHLORIDE | Erlotinib | Erotinib | N-(3-Ethynylphenyl)-6,7-bis(2-methoxyethoxy)-4-quinazolinamine Monohydrochloride | N-(3-ethynylphenyl)-6,7-bis(2-methoxyethoxy)quinazolin-4-amine | OSI-774 | Tarceva | US10189853, erlotinib | US10507209, Compound Erlotinib | US11524945, Compound Erlotinib | US9409845, Table 1, Compound 22: erlotinib | US9730934, Erlotinib | WO2022090481, Example erlotinib | cid_176870
Type:
Small organic molecule
Emp. Form.:
C22H23N3O4
Mol. Mass.:
393.4357
SMILES:
COCCOc1cc2ncnc(Nc3cccc(c3)C#C)c2cc1OCCOC