Reaction Details Report a problem with these data
Report a problem with these data
 Report a problem with these data
Report a problem with these dataTarget
3-hydroxy-3-methylglutaryl-coenzyme A reductase
Ligand
BDBM18372
Substrate
n/a
Meas. Tech.
ChEMBL_461936 (CHEMBL944783)
IC50
3.7±n/a nM
Citation
 Park, WK; Kennedy, RM; Larsen, SD; Miller, S; Roth, BD; Song, Y; Steinbaugh, BA; Sun, K; Tait, BD; Kowala, MC; Trivedi, BK; Auerbach, B; Askew, V; Dillon, L; Hanselman, JC; Lin, Z; Lu, GH; Robertson, A; Sekerke, C Hepatoselectivity of statins: design and synthesis of 4-sulfamoyl pyrroles as HMG-CoA reductase inhibitors. Bioorg Med Chem Lett 18:1151-6 (2008) [PubMed] Article
Park, WK; Kennedy, RM; Larsen, SD; Miller, S; Roth, BD; Song, Y; Steinbaugh, BA; Sun, K; Tait, BD; Kowala, MC; Trivedi, BK; Auerbach, B; Askew, V; Dillon, L; Hanselman, JC; Lin, Z; Lu, GH; Robertson, A; Sekerke, C Hepatoselectivity of statins: design and synthesis of 4-sulfamoyl pyrroles as HMG-CoA reductase inhibitors. Bioorg Med Chem Lett 18:1151-6 (2008) [PubMed] Article More Info.:
Target
Name:
3-hydroxy-3-methylglutaryl-coenzyme A reductase
Synonyms:
HMDH_RAT | HMG-CoA reductase | Hmgcr
Type:
Enzyme
Mol. Mass.:
96689.85
Organism:
Rattus norvegicus (rat)
Description:
Isolated rat liver microsomes were used as enzyme source.
Residue:
887
Sequence:
MLSRLFRMHGLFVASHPWEVIVGTVTLTICMMSMNMFTGNNKICGWNYECPKFEEDVLSSDIIILTITRCIAILYIYFQFQNLRQLGSKYILGIAGLFTIFSSFVFSTVVIHFLDKELTGLNEALPFFLLLIDLSRASALAKFALSSNSQDEVRENIARGMAILGPTFTLDALVECLVIGVGTMSGVRQLEIMCCFGCMSVLANYFVFMTFFPACVSLVLELSRESREGRPIWQLSHFARVLEEEENKPNPVTQRVKMIMSLGLVLVHAHSRWIADPSPQNSTAEQSKVSLGLAEDVSKRIEPSVSLWQFYLSKMISMDIEQVITLSLALLLAVKYIFFEQAETESTLSLKNPITSPVVTPKKAQDNCCRREPLLVRRNQKLSSVEEDPGVNQDRKVEVIKPLVAEAETSGRATFVLGASAASPPLALGAQEPGIELPSEPRPNEECLQILESAEKGAKFLSDAEIIQLVNAKHIPAYKLETLMETHERGVSIRRQLLSAKLAEPSSLQYLPYRDYNYSLVMGACCENVIGYMPIPVGVAGPLCLDGKEYQVPMATTEGCLVASTNRGCRAISLGGGASSRVLADGMSRGPVVRLPRACDSAEVKSWLETPEGFAVVKEAFDSTSRFARLQKLHVTLAGRNLYIRLQSKTGDAMGMNMISKGTEKALLKLQEFFPELQILAVSGNYCTDKKPAAINWIEGRGKTVVCEAVIPAKVVREVLKTTTEAMVDVNINKNLVGSAMAGSIGGYNAHAANIVTAIYIACGQDAAQNVGSSNCITLMEASGPTNEDLYISCTMPSIEIGTVGGGTNLLPQQACLQMLGVQGACKDNPGENARQLARIVCGTVMAGELSLMAALAAGHLVRSHMVHNRSKINLQDLQGTCTKKAA
Inhibitor
Name:
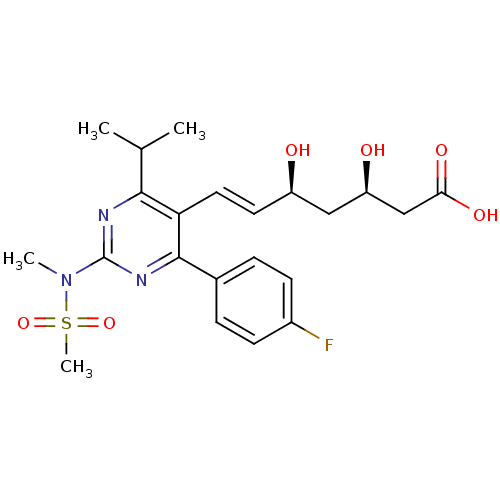
BDBM18372
Synonyms:
(3R,5S,6E)-7-[4-(4-fluorophenyl)-2-(N-methylmethanesulfonamido)-6-(propan-2-yl)pyrimidin-5-yl]-3,5-dihydroxyhept-6-enoic acid | CHEMBL1496 | Ros | Rosuvastatin | US9102656, Rosuvastatin | ZD4522
Type:
Small organic molecule
Emp. Form.:
C22H28FN3O6S
Mol. Mass.:
481.538
SMILES:
CC(C)c1nc(nc(-c2ccc(F)cc2)c1\C=C\[C@@H](O)C[C@@H](O)CC(O)=O)N(C)S(C)(=O)=O |r|