Reaction Details Report a problem with these data
Report a problem with these data
 Report a problem with these data
Report a problem with these dataTarget
Mitogen-activated protein kinase 8
Ligand
BDBM50277947
Substrate
n/a
Meas. Tech.
ChEMBL_501722 (CHEMBL985050)
Ki
9±n/a nM
Citation
 Humphries, PS; Lafontaine, JA; Agree, CS; Alexander, D; Chen, P; Do, QQ; Li, LY; Lunney, EA; Rajapakse, RJ; Siegel, K; Timofeevski, SL; Wang, T; Wilhite, DM Synthesis and SAR of 4-substituted-2-aminopyrimidines as novel c-Jun N-terminal kinase (JNK) inhibitors. Bioorg Med Chem Lett 19:2099-102 (2009) [PubMed] Article
Humphries, PS; Lafontaine, JA; Agree, CS; Alexander, D; Chen, P; Do, QQ; Li, LY; Lunney, EA; Rajapakse, RJ; Siegel, K; Timofeevski, SL; Wang, T; Wilhite, DM Synthesis and SAR of 4-substituted-2-aminopyrimidines as novel c-Jun N-terminal kinase (JNK) inhibitors. Bioorg Med Chem Lett 19:2099-102 (2009) [PubMed] Article More Info.:
Target
Name:
Mitogen-activated protein kinase 8
Synonyms:
JNK-46 | JNK1 | JNK1-alpha-1 | MAPK8 | MK08_HUMAN | Mitogen-Activated Protein Kinase 8 (JNK1) | PRKM8 | SAPK1 | SAPK1C | Stress-activated protein kinase JNK1 | c-Jun N-terminal kinase 1 | c-Jun N-terminal kinase 1 (JNK1) | c-Jun N-terminal kinase 1(JNK1) | c-Jun N-terminal kinase 2 (JNK2)
Type:
Enzyme
Mol. Mass.:
48297.57
Organism:
Homo sapiens (Human)
Description:
JNK-1 was purchased from Upstate Cell Signaling Solutions (formerly Upstate Biotechnology).
Residue:
427
Sequence:
MSRSKRDNNFYSVEIGDSTFTVLKRYQNLKPIGSGAQGIVCAAYDAILERNVAIKKLSRPFQNQTHAKRAYRELVLMKCVNHKNIIGLLNVFTPQKSLEEFQDVYIVMELMDANLCQVIQMELDHERMSYLLYQMLCGIKHLHSAGIIHRDLKPSNIVVKSDCTLKILDFGLARTAGTSFMMTPYVVTRYYRAPEVILGMGYKENVDLWSVGCIMGEMVCHKILFPGRDYIDQWNKVIEQLGTPCPEFMKKLQPTVRTYVENRPKYAGYSFEKLFPDVLFPADSEHNKLKASQARDLLSKMLVIDASKRISVDEALQHPYINVWYDPSEAEAPPPKIPDKQLDEREHTIEEWKELIYKEVMDLEERTKNGVIRGQPSPLGAAVINGSQHPSSSSSVNDVSSMSTDPTLASDTDSSLEAAAGPLGCCR
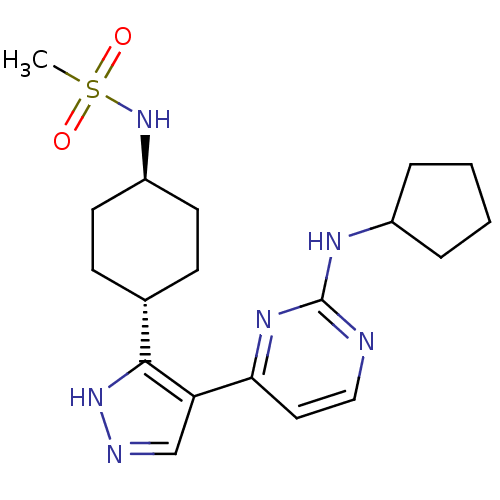
Inhibitor
Name:
BDBM50277947
Synonyms:
CHEMBL482559 | N-(4-(4-(2-(cyclopentylamino)pyrimidin-4-yl)-1H-pyrazol-3-yl)cyclohexyl)methanesulfonamide
Type:
Small organic molecule
Emp. Form.:
C19H28N6O2S
Mol. Mass.:
404.53
SMILES:
CS(=O)(=O)N[C@H]1CC[C@@H](CC1)c1[nH]ncc1-c1ccnc(NC2CCCC2)n1 |r,wU:8.11,wD:5.4,(22.41,-2.51,;22.42,-.98,;20.88,-.96,;23.96,-1,;22.44,.56,;23.79,1.31,;25.11,.52,;26.46,1.27,;26.48,2.81,;25.16,3.6,;23.82,2.85,;27.94,3.29,;28.41,4.76,;29.95,4.76,;30.43,3.3,;29.2,2.4,;29.2,.86,;27.87,.09,;27.87,-1.46,;29.21,-2.23,;30.54,-1.46,;31.88,-2.23,;31.88,-3.77,;30.64,-4.66,;31.11,-6.13,;32.66,-6.12,;33.13,-4.66,;30.54,.09,)|