Reaction Details Report a problem with these data
Report a problem with these data
 Report a problem with these data
Report a problem with these dataTarget
Prolyl endopeptidase FAP
Ligand
BDBM50338416
Substrate
n/a
Meas. Tech.
ChEMBL_728293 (CHEMBL1687450)
IC50
>35000±n/a nM
Citation
 Chen, P; Caldwell, CG; Ashton, W; Wu, JK; He, H; Lyons, KA; Thornberry, NA; Weber, AE Synthesis and evaluation of [(1R)-1-amino-2-(2,5-difluorophenyl)ethyl]cyclohexanes and 4-[(1R)-1-amino-2-(2,5-difluorophenyl)ethyl]piperidines as DPP-4 inhibitors. Bioorg Med Chem Lett 21:1880-6 (2011) [PubMed] Article
Chen, P; Caldwell, CG; Ashton, W; Wu, JK; He, H; Lyons, KA; Thornberry, NA; Weber, AE Synthesis and evaluation of [(1R)-1-amino-2-(2,5-difluorophenyl)ethyl]cyclohexanes and 4-[(1R)-1-amino-2-(2,5-difluorophenyl)ethyl]piperidines as DPP-4 inhibitors. Bioorg Med Chem Lett 21:1880-6 (2011) [PubMed] Article More Info.:
Target
Name:
Prolyl endopeptidase FAP
Synonyms:
170 kDa melanoma membrane-bound gelatinase | FAP | Fibroblast Activation Protein (FAP) | Fibroblast activation protein alpha | Integral membrane serine protease | SEPR_HUMAN | Seprase
Type:
Enzyme
Mol. Mass.:
87712.48
Organism:
Homo sapiens (Human)
Description:
Q12884
Residue:
760
Sequence:
MKTWVKIVFGVATSAVLALLVMCIVLRPSRVHNSEENTMRALTLKDILNGTFSYKTFFPNWISGQEYLHQSADNNIVLYNIETGQSYTILSNRTMKSVNASNYGLSPDRQFVYLESDYSKLWRYSYTATYYIYDLSNGEFVRGNELPRPIQYLCWSPVGSKLAYVYQNNIYLKQRPGDPPFQITFNGRENKIFNGIPDWVYEEEMLATKYALWWSPNGKFLAYAEFNDTDIPVIAYSYYGDEQYPRTINIPYPKAGAKNPVVRIFIIDTTYPAYVGPQEVPVPAMIASSDYYFSWLTWVTDERVCLQWLKRVQNVSVLSICDFREDWQTWDCPKTQEHIEESRTGWAGGFFVSTPVFSYDAISYYKIFSDKDGYKHIHYIKDTVENAIQITSGKWEAINIFRVTQDSLFYSSNEFEEYPGRRNIYRISIGSYPPSKKCVTCHLRKERCQYYTASFSDYAKYYALVCYGPGIPISTLHDGRTDQEIKILEENKELENALKNIQLPKEEIKKLEVDEITLWYKMILPPQFDRSKKYPLLIQVYGGPCSQSVRSVFAVNWISYLASKEGMVIALVDGRGTAFQGDKLLYAVYRKLGVYEVEDQITAVRKFIEMGFIDEKRIAIWGWSYGGYVSSLALASGTGLFKCGIAVAPVSSWEYYASVYTERFMGLPTKDDNLEHYKNSTVMARAEYFRNVDYLLIHGTADDNVHFQNSAQIAKALVNAQVDFQAMWYSDQNHGLSGLSTNHLYTHMTHFLKQCFSLSD
Inhibitor
Name:
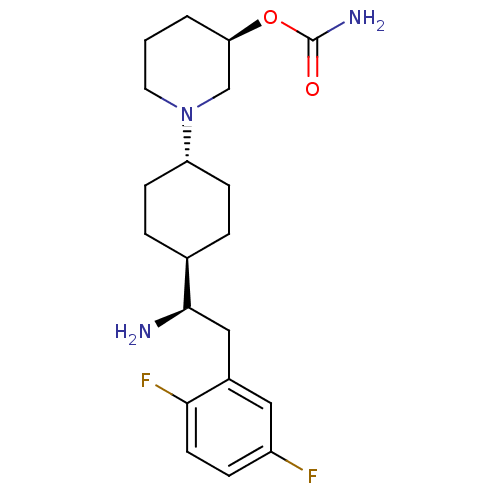
BDBM50338416
Synonyms:
CHEMBL1683122 | trans-(R)-1-(4-((R)-1-amino-2-(2,5-difluorophenyl)ethyl)cyclohexyl)piperidin-3-yl carbamate
Type:
Small organic molecule
Emp. Form.:
C20H29F2N3O2
Mol. Mass.:
381.46
SMILES:
N[C@H](Cc1cc(F)ccc1F)[C@H]1CC[C@@H](CC1)N1CCC[C@H](C1)OC(N)=O |r,wU:1.0,14.18,21.25,wD:11.17,(24.9,-6.17,;24.9,-7.71,;23.57,-8.49,;22.24,-7.72,;22.23,-6.17,;20.9,-5.4,;20.89,-3.86,;19.57,-6.18,;19.56,-7.72,;20.9,-8.49,;20.9,-10.03,;26.24,-8.48,;26.23,-10.02,;27.56,-10.79,;28.9,-10.02,;28.9,-8.48,;27.56,-7.7,;30.23,-10.79,;31.56,-10.02,;32.89,-10.8,;32.89,-12.34,;31.54,-13.1,;30.22,-12.33,;31.53,-14.64,;30.19,-15.4,;28.87,-14.63,;30.19,-16.94,)|