Reaction Details Report a problem with these data
Report a problem with these data
 Report a problem with these data
Report a problem with these dataTarget
MAP/microtubule affinity-regulating kinase 3
Ligand
BDBM50132260
Substrate
n/a
Meas. Tech.
ChEMBL_774343 (CHEMBL1908560)
Kd
>10000±n/a nM
Citation
 Davis, MI; Hunt, JP; Herrgard, S; Ciceri, P; Wodicka, LM; Pallares, G; Hocker, M; Treiber, DK; Zarrinkar, PP Comprehensive analysis of kinase inhibitor selectivity. Nat Biotechnol 29:1046-51 (2011) [PubMed] Article
Davis, MI; Hunt, JP; Herrgard, S; Ciceri, P; Wodicka, LM; Pallares, G; Hocker, M; Treiber, DK; Zarrinkar, PP Comprehensive analysis of kinase inhibitor selectivity. Nat Biotechnol 29:1046-51 (2011) [PubMed] Article More Info.:
Target
Name:
MAP/microtubule affinity-regulating kinase 3
Synonyms:
2.7.11.1 | C-TAK1 | CTAK1 | Cdc25C-associated protein kinase 1 | ELKL motif kinase 2 | EMK-2 | EMK2 | MAP/microtubule affinity-regulating kinase 3 | MAP/microtubule affinity-regulating kinase 3 (MARK 3) | MARK3 | MARK3_HUMAN | Par-1a | Protein kinase STK10 | Ser/Thr protein kinase PAR-1 | Serine/threonine-protein kinase c-TAK1 | Serine/threonine-protein kinase p78
Type:
Protein
Mol. Mass.:
84525.94
Organism:
Homo sapiens (Human)
Description:
P27448
Residue:
753
Sequence:
MSTRTPLPTVNERDTENHTSHGDGRQEVTSRTSRSGARCRNSIASCADEQPHIGNYRLLKTIGKGNFAKVKLARHILTGREVAIKIIDKTQLNPTSLQKLFREVRIMKILNHPNIVKLFEVIETEKTLYLIMEYASGGEVFDYLVAHGRMKEKEARSKFRQIVSAVQYCHQKRIVHRDLKAENLLLDADMNIKIADFGFSNEFTVGGKLDTFCGSPPYAAPELFQGKKYDGPEVDVWSLGVILYTLVSGSLPFDGQNLKELRERVLRGKYRIPFYMSTDCENLLKRFLVLNPIKRGTLEQIMKDRWINAGHEEDELKPFVEPELDISDQKRIDIMVGMGYSQEEIQESLSKMKYDEITATYLLLGRKSSELDASDSSSSSNLSLAKVRPSSDLNNSTGQSPHHKVQRSVSSSQKQRRYSDHAGPAIPSVVAYPKRSQTSTADSDLKEDGISSRKSSGSAVGGKGIAPASPMLGNASNPNKADIPERKKSSTVPSSNTASGGMTRRNTYVCSERTTADRHSVIQNGKENSTIPDQRTPVASTHSISSAATPDRIRFPRGTASRSTFHGQPRERRTATYNGPPASPSLSHEATPLSQTRSRGSTNLFSKLTSKLTRRNMSFRFIKRLPTEYERNGRYEGSSRNVSAEQKDENKEAKPRSLRFTWSMKTTSSMDPGDMMREIRKVLDANNCDYEQRERFLLFCVHGDGHAENLVQWEMEVCKLPRLSLNGVRFKRISGTSIAFKNIASKIANELKL
Inhibitor
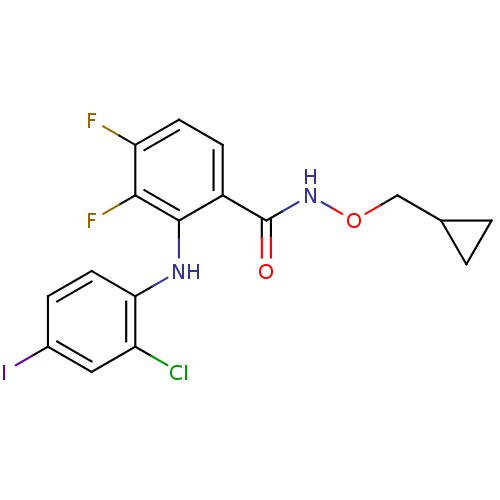
Name:
BDBM50132260
Synonyms:
2-(2-Chloro-4-iodo-phenylamino)-N-cyclopropylmethoxy-3,4-difluoro-benzamide | 2-(2-chloro-4-iodophenylamino)-N-(cyclopropylmethoxy)-3,4-difluorobenzamide | CHEMBL105442 | CI-1040 | PD-18435 | PD-184352 | US8575391, 341
Type:
Small organic molecule
Emp. Form.:
C17H14ClF2IN2O2
Mol. Mass.:
478.66
SMILES:
Fc1ccc(C(=O)NOCC2CC2)c(Nc2ccc(I)cc2Cl)c1F