Reaction Details Report a problem with these data
Report a problem with these data
 Report a problem with these data
Report a problem with these dataTarget
Orotidine 5'-phosphate decarboxylase
Ligand
BDBM21340
Substrate
n/a
Meas. Tech.
ChEMBL_381850 (CHEMBL869172)
Ki
64±n/a nM
Citation
More Info.:
Target
Name:
Orotidine 5'-phosphate decarboxylase
Synonyms:
Orotidine phosphate decarboxylase | PYRF_YEAST | URA3
Type:
PROTEIN
Mol. Mass.:
29241.18
Organism:
Saccharomyces cerevisiae
Description:
ChEMBL_639015
Residue:
267
Sequence:
MSKATYKERAATHPSPVAAKLFNIMHEKQTNLCASLDVRTTKELLELVEALGPKICLLKTHVDILTDFSMEGTVKPLKALSAKYNFLLFEDRKFADIGNTVKLQYSAGVYRIAEWADITNAHGVVGPGIVSGLKQAAEEVTKEPRGLLMLAELSCKGSLATGEYTKGTVDIAKSDKDFVIGFIAQRDMGGRDEGYDWLIMTPGVGLDDKGDALGQQYRTVDDVVSTGSDIIIVGRGLFAKGRDAKVEGERYRKAGWEAYLRRCGQQN
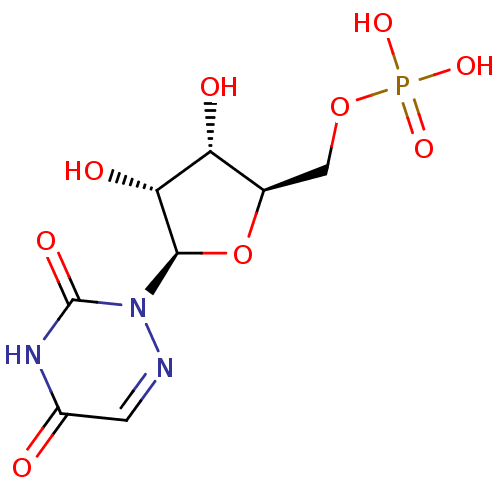
Inhibitor
Name:
BDBM21340
Synonyms:
6-aza-UMP | C6-Uridine Derivative, 18 | {[(2R,3S,4R,5R)-5-(3,5-dioxo-2,3,4,5-tetrahydro-1,2,4-triazin-2-yl)-3,4-dihydroxyoxolan-2-yl]methoxy}phosphonic acid
Type:
Small organic molecule
Emp. Form.:
C8H12N3O9P
Mol. Mass.:
325.1693
SMILES:
O[C@@H]1[C@@H](COP(O)(O)=O)O[C@H]([C@@H]1O)n1ncc(=O)[nH]c1=O |r|