Reaction Details Report a problem with these data
Report a problem with these data
 Report a problem with these data
Report a problem with these dataTarget
Peptide deformylase 1A, chloroplastic/mitochondrial
Ligand
BDBM50201883
Substrate
n/a
Meas. Tech.
ChEMBL_423228 (CHEMBL912247)
IC50
>1000000±n/a nM
Citation
 Boularot, A; Giglione, C; Petit, S; Duroc, Y; Alves de Sousa, R; Larue, V; Cresteil, T; Dardel, F; Artaud, I; Meinnel, T Discovery and refinement of a new structural class of potent peptide deformylase inhibitors. J Med Chem 50:10-20 (2007) [PubMed] Article
Boularot, A; Giglione, C; Petit, S; Duroc, Y; Alves de Sousa, R; Larue, V; Cresteil, T; Dardel, F; Artaud, I; Meinnel, T Discovery and refinement of a new structural class of potent peptide deformylase inhibitors. J Med Chem 50:10-20 (2007) [PubMed] Article More Info.:
Target
Name:
Peptide deformylase 1A, chloroplastic/mitochondrial
Synonyms:
AtDEF1 | AtDEF1A | DEF1 | DEF1A_ARATH | PDF 1A | PDF1A | Peptide deformylase 1A, chloroplastic | Polypeptide deformylase
Type:
PROTEIN
Mol. Mass.:
30001.50
Organism:
Arabidopsis thaliana
Description:
ChEMBL_423228
Residue:
269
Sequence:
MGLHRDEATAMETLFRVSLRLLPVSAAVTCRSIRFPVSRPGSSHLLNRKLYNLPTSSSSSLSTKAGWLLGLGEKKKKVDLPEIVASGDPVLHEKAREVDPGEIGSERIQKIIDDMIKVMRLAPGVGLAAPQIGVPLRIIVLEDTKEYISYAPKEEILAQERRHFDLMVMVNPVLKERSNKKALFFEGCLSVDGFRAAVERYLEVVVTGYDRQGKRIEVNASGWQARILQHECDHLDGNLYVDKMVPRTFRTVDNLDLPLAEGCPKLGPQ
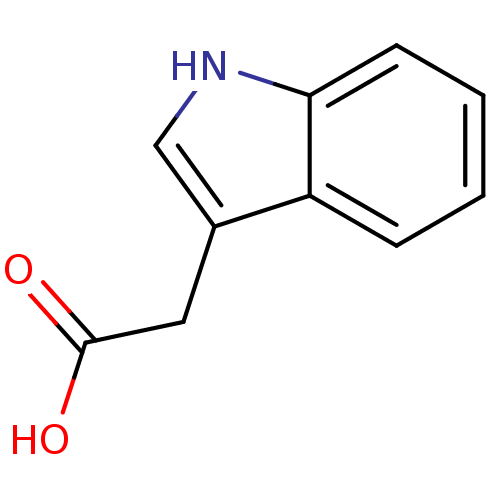
Inhibitor
Name:
BDBM50201883
Synonyms:
(indol-3-yl)acetic acid | 1H-indol-3-ylacetic acid | 2-(indol-3-yl)ethanoic acid | 3-Indolylessigsaeure | Acid, 6 | CHEMBL82411 | IAA | IES | heteroauxin | indole-3-acetic acid
Type:
Small organic molecule
Emp. Form.:
C10H9NO2
Mol. Mass.:
175.184
SMILES:
OC(=O)Cc1c[nH]c2ccccc12