Reaction Details Report a problem with these data
Report a problem with these data
 Report a problem with these data
Report a problem with these dataTarget
Serine protease hepsin
Ligand
BDBM50286441
Substrate
n/a
Meas. Tech.
ChEMBL_1432692 (CHEMBL3389936)
Ki
61±n/a nM
Citation
 Han, Z; Harris, PK; Jones, DE; Chugani, R; Kim, T; Agarwal, M; Shen, W; Wildman, SA; Janetka, JW Inhibitors of HGFA, Matriptase, and Hepsin Serine Proteases: A Nonkinase Strategy to Block Cell Signaling in Cancer. ACS Med Chem Lett 5:1219-24 (2014) [PubMed] Article
Han, Z; Harris, PK; Jones, DE; Chugani, R; Kim, T; Agarwal, M; Shen, W; Wildman, SA; Janetka, JW Inhibitors of HGFA, Matriptase, and Hepsin Serine Proteases: A Nonkinase Strategy to Block Cell Signaling in Cancer. ACS Med Chem Lett 5:1219-24 (2014) [PubMed] Article More Info.:
Target
Name:
Serine protease hepsin
Synonyms:
HEPS_HUMAN | HPN | Hepsin | Serine protease hepsin | TMPRSS1 | Transmembrane protease serine 11E | Transmembrane protease, serine 1
Type:
Enzyme
Mol. Mass.:
45016.98
Organism:
Homo sapiens (Human)
Description:
Human deglycosylated hepsin was expressed in Pichia pastoris.
Residue:
417
Sequence:
MAQKEGGRTVPCCSRPKVAALTAGTLLLLTAIGAASWAIVAVLLRSDQEPLYPVQVSSADARLMVFDKTEGTWRLLCSSRSNARVAGLSCEEMGFLRALTHSELDVRTAGANGTSGFFCVDEGRLPHTQRLLEVISVCDCPRGRFLAAICQDCGRRKLPVDRIVGGRDTSLGRWPWQVSLRYDGAHLCGGSLLSGDWVLTAAHCFPERNRVLSRWRVFAGAVAQASPHGLQLGVQAVVYHGGYLPFRDPNSEENSNDIALVHLSSPLPLTEYIQPVCLPAAGQALVDGKICTVTGWGNTQYYGQQAGVLQEARVPIISNDVCNGADFYGNQIKPKMFCAGYPEGGIDACQGDSGGPFVCEDSISRTPRWRLCGIVSWGTGCALAQKPGVYTKVSDFREWIFQAIKTHSEASGMVTQL
Inhibitor
Name:
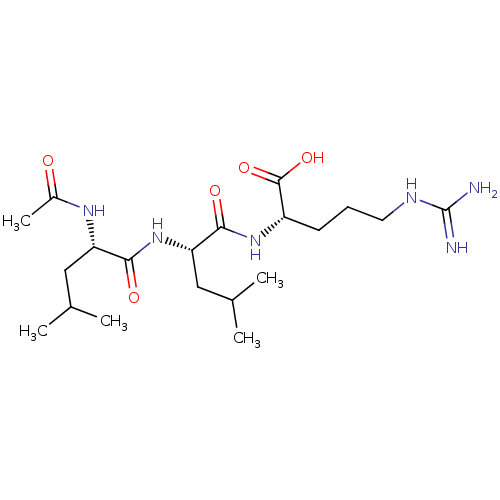
BDBM50286441
Synonyms:
(S)-2-((S)-2-Acetylamino-4-(S)-methyl-pentanoylamino)-4-methyl-pentanoic acid (1-formyl-4-guanidino-butyl)-amide | (S)-2-Acetylamino-4-methyl-pentanoic acid [(R)-1-((S)-1-formyl-4-guanidino-butylcarbamoyl)-3-methyl-butyl]-amide | (S)-2-Acetylamino-4-methyl-pentanoic acid [(S)-1-((S)-1-formyl-4-guanidino-butylcarbamoyl)-3-methyl-butyl]-amide | (S)-2-[(S)-2-((S)-2-Acetylamino-4-methyl-pentanoylamino)-4-methyl-pentanoylamino]-5-guanidino-pentanoic acid | (S)-2-acetylamino-4-methyl-pentanoic acid [(S)-2-(1-formyl-4-guanidino-butylamino)-4-methyl-pentanoyl]-amide | 2-Acetylamino-4-methyl-pentanoic acid [1-(1-formyl-4-guanidino-butylcarbamoyl)-3-methyl-butyl]-amide | Ac-LLR-CHO | Ac-Leu-Leu-Arg-H | CHEMBL129157
Type:
Small organic molecule
Emp. Form.:
C20H38N6O5
Mol. Mass.:
442.5529
SMILES:
CC(C)C[C@H](NC(C)=O)C(=O)N[C@@H](CC(C)C)C(=O)N[C@@H](CCCNC(N)=N)C(O)=O