Reaction Details Report a problem with these data
Report a problem with these data
 Report a problem with these data
Report a problem with these dataTarget
Lysosomal acid glucosylceramidase
Ligand
BDBM50196633
Substrate
n/a
Meas. Tech.
ChEMBL_1618764 (CHEMBL3860933)
pH
5.9±n/a
IC50
251±n/a nM
Comments
extracted
Citation
 Zheng, J; Chen, L; Schwake, M; Silverman, RB; Krainc, D Design and Synthesis of Potent Quinazolines as Selective▀-Glucocerebrosidase Modulators. J Med Chem 59:8508-20 (2016) [PubMed] Article
Zheng, J; Chen, L; Schwake, M; Silverman, RB; Krainc, D Design and Synthesis of Potent Quinazolines as Selective▀-Glucocerebrosidase Modulators. J Med Chem 59:8508-20 (2016) [PubMed] Article More Info.:
Target
Name:
Lysosomal acid glucosylceramidase
Synonyms:
Acid beta-glucosidase | Alglucerase | Beta-glucocerebrosidase | Beta-glucocerebrosidase (GC) | D-glucosyl-N-acylsphingosine glucohydrolase | GBA | GBA1 | GBA1_HUMAN | GC | GCase | GLUC | Glucocerebrosidase (GBA) | Glucosylceramidase (GBA) | Glucosylceramidase (GCase) | Glucosylceramidase precursor (Beta-glucocerebrosidase) (Acid beta-glucosidase) (D-glucosyl-N-acylsphingosine glucohydrolase) (Alglucerase) (Imiglucerase) | Imiglucerase | beta-glucocerebrosidase (GCase)
Type:
Enzyme
Mol. Mass.:
59724.64
Organism:
Homo sapiens (Human)
Description:
The beta-Glu activity was measured with commercially available beta-glucocerebrosidase (Ceredase) as the enzyme source.
Residue:
536
Sequence:
MEFSSPSREECPKPLSRVSIMAGSLTGLLLLQAVSWASGARPCIPKSFGYSSVVCVCNATYCDSFDPPTFPALGTFSRYESTRSGRRMELSMGPIQANHTGTGLLLTLQPEQKFQKVKGFGGAMTDAAALNILALSPPAQNLLLKSYFSEEGIGYNIIRVPMASCDFSIRTYTYADTPDDFQLHNFSLPEEDTKLKIPLIHRALQLAQRPVSLLASPWTSPTWLKTNGAVNGKGSLKGQPGDIYHQTWARYFVKFLDAYAEHKLQFWAVTAENEPSAGLLSGYPFQCLGFTPEHQRDFIARDLGPTLANSTHHNVRLLMLDDQRLLLPHWAKVVLTDPEAAKYVHGIAVHWYLDFLAPAKATLGETHRLFPNTMLFASEACVGSKFWEQSVRLGSWDRGMQYSHSIITNLLYHVVGWTDWNLALNPEGGPNWVRNFVDSPIIVDITKDTFYKQPMFYHLGHFSKFIPEGSQRVGLVASQKNDLDAVALMHPDGSAVVVVLNRSSKDVPLTIKDPAVGFLETISPGYSIHTYLWRRQ
Inhibitor
Name:
BDBM50196633
Synonyms:
CHEMBL3965871 | US10167270, Compound 6d
Type:
Small organic molecule
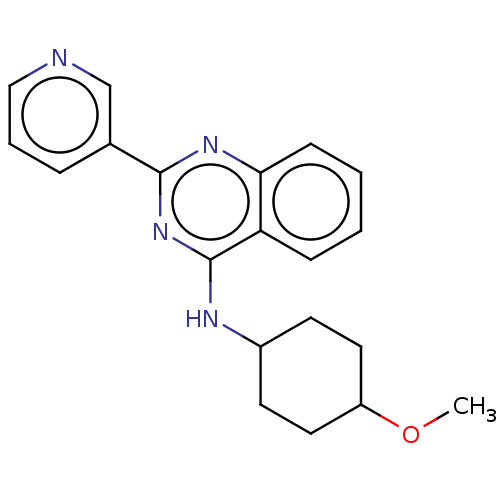
Emp. Form.:
C20H22N4O
Mol. Mass.:
334.4149
SMILES:
COC1CCC(CC1)Nc1nc(nc2ccccc12)-c1cccnc1 |(62.32,-14.11,;63.65,-14.88,;64.99,-14.12,;64.99,-12.58,;66.32,-11.81,;67.65,-12.57,;67.66,-14.12,;66.32,-14.89,;68.98,-11.8,;68.98,-10.26,;70.32,-9.49,;70.31,-7.94,;68.97,-7.18,;67.64,-7.95,;66.31,-7.19,;64.98,-7.96,;64.98,-9.5,;66.31,-10.27,;67.65,-9.49,;71.64,-7.16,;72.97,-7.93,;74.3,-7.15,;74.29,-5.61,;72.94,-4.85,;71.62,-5.63,)|