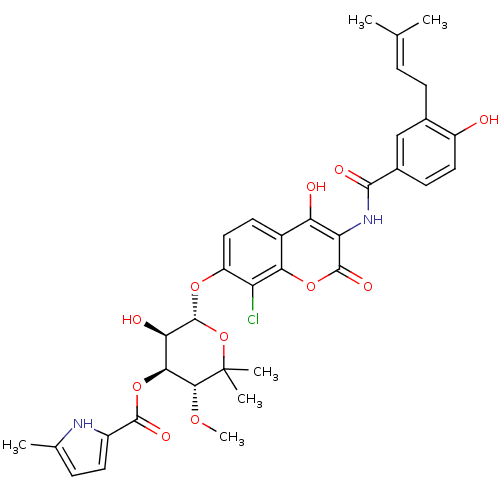
Reaction Details Report a problem with these data
Report a problem with these data
 Report a problem with these data
Report a problem with these dataTarget
DNA gyrase subunit A
Ligand
BDBM50330317
Substrate
n/a
Meas. Tech.
ChEMBL_1636458 (CHEMBL3879356)
IC50
929±n/a nM
Citation
 Alagumuthu, M; Arumugam, S Molecular docking, discovery, synthesis, and pharmacological properties of new 6-substituted-2-(3-phenoxyphenyl)-4-phenyl quinoline derivatives; an approach to developing potent DNA gyrase inhibitors/antibacterial agents. Bioorg Med Chem 25:1448-1455 (2017) [PubMed] Article
Alagumuthu, M; Arumugam, S Molecular docking, discovery, synthesis, and pharmacological properties of new 6-substituted-2-(3-phenoxyphenyl)-4-phenyl quinoline derivatives; an approach to developing potent DNA gyrase inhibitors/antibacterial agents. Bioorg Med Chem 25:1448-1455 (2017) [PubMed] Article More Info.:
Target
Name:
DNA gyrase subunit A
Synonyms:
DNA gyrase subunit A (gyrA) | GYRA_STAAU | gyrA
Type:
Enzyme Subunit
Mol. Mass.:
99588.82
Organism:
Staphylococcus aureus
Description:
n/a
Residue:
889
Sequence:
MAELPQSRINERNITSEMRESFLDYAMSVIVARALPDVRDGLKPVHRRILYGLNEQGMTPDKSYKKSARIVGDVMGKYHPHGDSSIYEAMVRMAQDFSYRYPLVDGQGNFGSMDGDGAAAMRYTEARMTKITLELLRDINKDTIDFIDNYDGNEREPSVLPARFPNLLANGASGIAVGMATNIPPHNLTELINGVLSLSKNPDISIAELMEDIEGPDFPTAGLILGKSGIRRAYETGRGSIQMRSRAVIEERGGGRQRIVVTEIPFQVNKARMIEKIAELVRDKKIDGITDLRDETSLRTGVRVVIDVRKDANASVILNNLYKQTPLQTSFGVNMIALVNGRPKLINLKEALVHYLEHQKTVVRRRTQYNLRKAKDRAHILEGLRIALDHIDEIISTIRESDTDKVAMESLQQRFKLSEKQAQAILDMRLRRLTGLERDKIEAEYNELLNYISELEAILADEEVLLQLVRDELTEIRDRFGDDRRTEIQLGGFEDLEDEDLIPEEQIVITLSHNNYIKRLPVSTYRAQNRGGRGVQGMNTLEEDFVSQLVTLSTHDHVLFFTNKGRVYKLKGYEVPELSRQSKGIPVVNAIELENDEVISTMIAVKDLESEDNFLVFATKRGVVKRSALSNFSRINRNGKIAISFREDDELIAVRLTSGQEDILIGTSHASLIRFPESTLRPLGRTATGVKGITLREGDEVVGLDVAHANSVDEVLVVTENGYGKRTPVNDYRLSNRGGKGIKTATITERNGNVVCITTVTGEEDLMIVTNAGVIIRLDVADISQNGRAAQGVRLIRLGDDQFVSTVAKVKEDAEDETNEDEQSTSTVSEDGTEQQREAVVNDETPGNAIHTEVIDSEENDEDGRIEVRQDFMDRVEEDIQQSLDEDEE
Inhibitor
Name:
BDBM50330317
Synonyms:
5-Methyl-1H-pyrrole-2-carboxylic acid (3R,4S,5R,6S)-6-{8-chloro-4-hydroxy-3-[4-hydroxy-3-(3-methyl-but-2-enyl)-benzoylamino]-2-oxo-2H-chromen-7-yloxy}-5-hydroxy-3-methoxy-2,2-dimethyl-tetrahydro-pyran-4-yl ester | 5-Methyl-1H-pyrrole-2-carboxylic acid (3R,4S,5R,6S)-6-{8-chloro-4-hydroxy-3-[4-hydroxy-3-(4-methyl-pent-3-enyl)-benzoylamino]-2-oxo-2H-chromen-7-yloxy}-5-hydroxy-3-methoxy-2,2-dimethyl-tetrahydro-pyran-4-yl ester | CHEMBL303984 | Clorobiocin
Type:
Small organic molecule
Emp. Form.:
C35H37ClN2O11
Mol. Mass.:
697.128
SMILES:
CO[C@@H]1[C@@H](OC(=O)c2ccc(C)[nH]2)[C@@H](O)[C@H](Oc2ccc3c(O)c(NC(=O)c4ccc(O)c(CC=C(C)C)c4)c(=O)oc3c2Cl)OC1(C)C |r,wU:3.3,13.14,wD:2.1,15.16,(-8.34,-2.73,;-8.34,-1.19,;-7.01,-.42,;-7.01,1.12,;-8.34,1.89,;-9.68,1.12,;-9.68,-.42,;-11.01,1.89,;-11.17,3.42,;-12.68,3.74,;-13.45,2.4,;-14.98,2.24,;-12.42,1.26,;-5.68,1.89,;-5.68,3.43,;-4.34,1.12,;-3.01,1.89,;-1.67,1.12,;-1.67,-.42,;-.34,-1.19,;.99,-.42,;2.33,-1.19,;2.33,-2.73,;3.66,-.42,;4.99,-1.19,;6.33,-.42,;6.33,1.12,;7.66,-1.19,;7.66,-2.73,;8.99,-3.5,;10.33,-2.73,;11.66,-3.5,;10.33,-1.19,;11.66,-.42,;13,-1.19,;14.33,-.42,;15.66,-1.19,;14.33,1.12,;8.99,-.42,;3.66,1.12,;4.99,1.89,;2.33,1.89,;.99,1.12,;-.34,1.89,;-.34,3.43,;-4.34,-.42,;-5.68,-1.19,;-6.67,-2.37,;-4.69,-2.37,)|