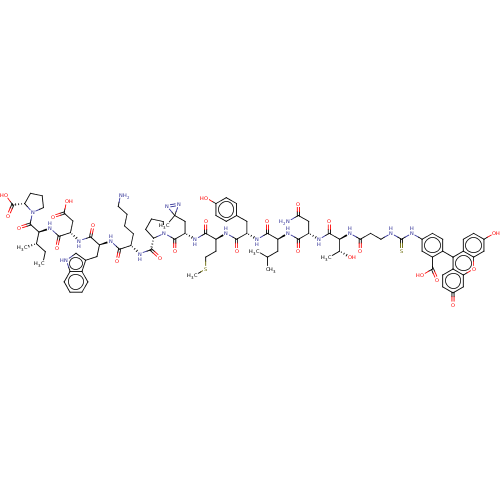
Reaction Details Report a problem with these data
Report a problem with these data
 Report a problem with these data
Report a problem with these dataTarget
UDP-3-O-(3-hydroxymyristoyl)glucosamine N-acyltransferase
Ligand
BDBM228808
Substrate
n/a
Meas. Tech.
Fluorescence Polarization Assay
pH
8±n/a
Kd
1.1e+3± 1e+2 nM
Comments
extracted
Citation
 Jenkins, RJ; Heslip, KA; Meagher, JL; Stuckey, JA; Dotson, GD Structural basis for the recognition of peptide RJPXD33 by acyltransferases in lipid A biosynthesis. J Biol Chem 289:15527-35 (2014) [PubMed] Article
Jenkins, RJ; Heslip, KA; Meagher, JL; Stuckey, JA; Dotson, GD Structural basis for the recognition of peptide RJPXD33 by acyltransferases in lipid A biosynthesis. J Biol Chem 289:15527-35 (2014) [PubMed] Article More Info.:
Target
Name:
UDP-3-O-(3-hydroxymyristoyl)glucosamine N-acyltransferase
Synonyms:
LPXD_ECOLI | UDP-3-O-(acryl)-glucosamine acryltransferase (LpxD) | firA | lpxD | omsA
Type:
Enzyme
Mol. Mass.:
36038.11
Organism:
Escherichia coli (Enterobacteria)
Description:
P21645
Residue:
341
Sequence:
MPSIRLADLAQQLDAELHGDGDIVITGVASMQSAQTGHITFMVNPKYREHLGLCQASAVVMTQDDLPFAKSAALVVKNPYLTYARMAQILDTTPQPAQNIAPSAVIDATAKLGNNVSIGANAVIESGVELGDNVIIGAGCFVGKNSKIGAGSRLWANVTIYHEIQIGQNCLIQSGTVVGADGFGYANDRGNWVKIPQIGRVIIGDRVEIGACTTIDRGALDDTIIGNGVIIDNQCQIAHNVVIGDNTAVAGGVIMAGSLKIGRYCMIGGASVINGHMEICDKVTVTGMGMVMRPITEPGVYSSGIPLQPNKVWRKTAALVMNIDDMSKRLKSLERKVNQQD
Inhibitor
Name:
BDBM228808
Synonyms:
FITC-(βa)TNLYM(L-photoleucine)PKWDIP-CONH2 | FITC-Photo 3
Type:
Small organic molecule
Emp. Form.:
C94H119N19O24S2
Mol. Mass.:
1963.194
SMILES:
CC[C@H](C)[C@H](NC(=O)[C@H](CC(O)=O)NC(=O)[C@H](Cc1c[nH]c2ccccc12)NC(=O)[C@H](CCCCN)NC(=O)[C@@H]1CCCN1C(=O)[C@H](CC1(C)N=N1)NC(=O)[C@H](CCSC)NC(=O)[C@H](Cc1ccc(O)cc1)NC(=O)[C@H](CC(C)C)NC(=O)[C@H](CC(N)=O)NC(=O)[C@@H](NC(=O)CCNC(=S)Nc1ccc(c(c1)C(O)=O)-c1c2ccc(O)cc2oc2cc(=O)ccc12)[C@@H](C)O)C(=O)N1CCC[C@H]1C(O)=O |r,wU:126.136,75.84,55.63,91.96,39.40,16.28,4.4,wD:83.92,63.76,46.55,30.37,8.12,2.2,135.145,c:53,(58.94,-19.08,;58.41,-20.52,;59.4,-21.7,;60.92,-21.43,;58.87,-23.15,;57.36,-23.42,;56.17,-22.17,;56.7,-20.72,;54.66,-22.43,;54.13,-23.88,;55.12,-25.06,;56.64,-24.79,;54.59,-26.51,;53.67,-21.25,;51.96,-21.45,;51.43,-22.9,;50.97,-20.27,;51.49,-18.82,;53.01,-18.56,;54.11,-19.62,;55.46,-18.9,;55.2,-17.39,;56.15,-16.18,;55.58,-14.76,;54.07,-14.54,;53.12,-15.74,;53.68,-17.17,;49.45,-20.54,;48.27,-19.29,;48.79,-17.84,;46.75,-19.56,;46.22,-21,;47.21,-22.18,;46.69,-23.63,;47.68,-24.81,;47.15,-26.26,;45.76,-18.38,;44.06,-18.54,;43.53,-19.99,;43.01,-17.41,;43.3,-15.9,;41.96,-15.16,;40.83,-16.21,;41.48,-17.6,;40.14,-16.91,;40.14,-15.37,;38.81,-17.68,;38.81,-19.22,;40.14,-19.99,;41.48,-19.22,;39.37,-21.32,;40.91,-21.32,;37.48,-16.91,;36.14,-17.68,;36.14,-19.22,;34.81,-16.91,;34.81,-15.37,;36.14,-14.6,;36.14,-13.06,;37.48,-12.29,;33.48,-17.68,;31.94,-16.91,;31.94,-15.37,;30.6,-17.68,;30.6,-19.22,;31.94,-19.99,;31.94,-21.53,;33.27,-22.3,;34.6,-21.53,;35.94,-22.3,;34.6,-19.99,;33.27,-19.22,;29.27,-16.91,;27.73,-17.68,;27.73,-19.22,;26.4,-16.91,;26.4,-15.37,;27.73,-14.6,;29.06,-15.37,;27.73,-13.06,;25.06,-17.68,;23.52,-16.91,;23.52,-15.37,;22.19,-17.68,;22.19,-19.22,;23.52,-19.99,;23.52,-21.53,;24.86,-19.22,;20.85,-16.91,;19.31,-17.68,;19.31,-19.22,;17.98,-16.91,;16.64,-17.68,;15.43,-16.91,;15.43,-15.37,;14.1,-17.68,;12.76,-16.91,;11.43,-17.68,;10.1,-16.91,;10.1,-15.37,;8.76,-17.68,;8.76,-19.22,;10.1,-19.99,;10.1,-21.53,;8.76,-22.3,;7.43,-21.53,;7.43,-19.99,;5.94,-21.93,;4.85,-20.84,;5.54,-23.42,;8.76,-23.84,;10.1,-24.61,;11.43,-23.84,;12.76,-24.61,;12.76,-26.15,;14.1,-26.92,;11.43,-26.92,;10.1,-26.15,;8.76,-26.92,;7.43,-26.15,;6.1,-26.92,;4.76,-26.15,;3.43,-26.92,;4.76,-24.61,;6.1,-23.84,;7.43,-24.61,;17.98,-15.37,;16.65,-14.6,;19.31,-14.6,;59.86,-24.33,;59.34,-25.78,;61.54,-24.16,;60.15,-24.81,;60.34,-26.34,;61.85,-26.63,;62.59,-25.29,;64.12,-25.1,;65.08,-26.3,;64.65,-23.65,)|